Introduction
Hanwoo cattle (HC) are Zebu type or (Bos indicus); characterized by small-bodied, slow maturity, good adaptability, excellent flavor and marbling meat (Song, 1994). They are considered indigenous to South Korea and are distinguished into four breeds with different colors, namely brown, brindle, black, and Jeju black Hanwoo (Figure 1). Among the breeds, the brown Hanwoo is the most popular breed (Lee et al., 2014). Years back, Hanwoo cattle were used for farming, transportation and religious activities, but for the past 30 years, they are developed into a meat-type breed. The estimated population of HC is about 3.2 million head in the fourth quarter of 2019 (KOSIS, 2019). Their contribution to beef meat is about 30% among the carcass meat produced in the country (KOSIS, 2017).
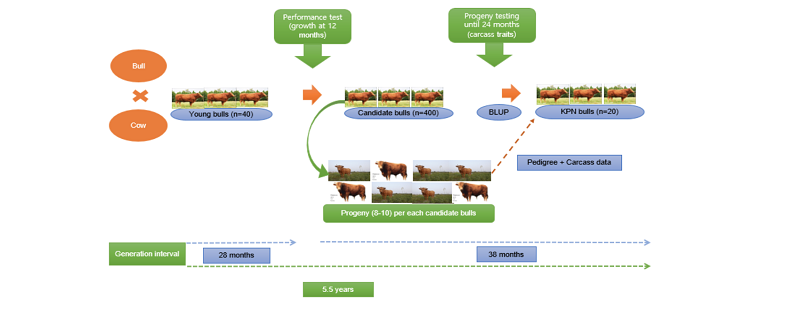
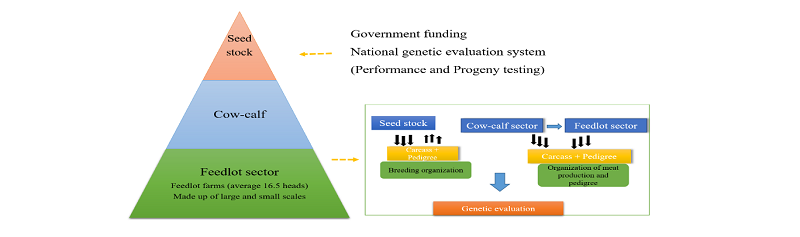
Genetic improvement provides a significant technique in which the cattle producers can improve the performance of their herds. It further comprises selection of superior animals from a population to produce better yields in the next generations. Genetic improvement also takes place when a genetic merit of an individual is upgraded through the selection of breeding objective or economically important traits. These selected economic traits contribute to the overall breeding objective, such as carcass yield or high growth rate (Sonstegard et al., 2001). In general, the genetic improvement in HC was set-up through the establishment of breeding programs, namely Hanwoo-Gaeryang-Danji (HGD) and Hanwoo-Gaeryang-Nongga (HGN) within the country (MAF, 1999). The production system also was designed towards achieving the breeding objectives. It comprises a three-tier system, namely the seed stock, the cow-calf and the feedlots. The breeding and production methods encourage the transfer of genetic merit from one stage to another. The methods further provide huge information such as performance, progeny test, and pedigree records used for estimation of genetic parameters and genetic improvement of the animals (Figures 2 & 3).
As it stands, the practice of animal recording by the national and private services has aided in the genetic evaluation. At the same time, the results of evaluations using pedigree records and phenotypic performance of individuals have shown the rate of genetic improvement (Song, 1994; Yoon et al., 2002; Park et al., 2013). The use of animal records has supported the selection of superior animals through their estimated breeding values (EBVs). In this regard, the assessment of EBVs and genetic parameters in HC is important to measure the trend of genetic merit. On the other hand, applying genomic selection is a potential approach to improve the genetic gains in economically important traits. Genomic selection is a useful tool for selecting traits with low heritability, difficult and expensive to measure (Meuwissen et al., 2016). A high prediction accuracy of genomic estimated breeding value (GEBV) encourages selection accuracy; increases genetic gain and reduces the generation interval in dairy cattle (Hayes et al., 2009; VanRaden et al., 2009; Harris and Johnson, 2010; Su et al., 2012). Previous studies have reviewed the breeding initiatives for HC to thrive as a beef industry (Sidong et al., 2017) and development of breeding and feeding systems (Chung et al., 2018). Therefore, the present study reviews the genetic improvement and application of genomic selection in Korean Hanwoo cattle.
Historical overview of Korean Hanwoo cattle
The origin of HC has drawn the attention of many researchers and studies. Based on the archaeological findings, HC has been reported to originate from crosses between Bos primigenius and Bos Zebu. The crossing between the ancestors was assumed to occur in Northern China and Mongolia. As a result, the ancestors of the HC appeared to migrate to the Korean peninsula in the New Stone Age (Song, 1994). A report by McTavish et al. (2013) also revealed that Asian cattle were crossbred of both taurine and indicine cattle. Based on diversity study among cattle breeds, it reveals that Asia cattle is different from Western taurine cattle (Lee et al., 2014). In all cases, the studies by McTavish et al. (2013) and Lee et al. (2014) are comparable with the previous report by Mannen et al. (2004), who analyzed a data comprised of African, European, Asian taurine and zebu cattle. In his findings, it shows that the Hanwoo tree was separated from the European type.
Breeding program, objective and production system
Hanwoo cattle possess a good meat flavor, tenderness, taste; however, with low productive performance compared with some breeds, and efforts have been made to improve the quantity and quality of the carcass. In the 1970s, the genetic improvement of HC started on a small-scale, but the program was unsuccessful. In the mid-80s, a new approach for an extensive breeding program for improvement of HC became a reality. The breeding program involves selection, design mating and testing procedures (Song, 1994). Lee et al. (2013) explained the breeding program of HC. The authors describe two initiatives, namely HGD created in 1979 by the Ministry of Agriculture and Forestry (MAF) and HGN in 1999, respectively. The area of coverage for HGD program is within a specific province, though; its purpose was not fully achieved due to ineffectiveness or shortage of competent workforce to take appropriate records. In contrast, the HGN program was created to focus on individual farms. During this shift in the program, it reveals that the parents of HGN were from the HGD program, and this process encourages proper evaluation and testing of animals’ performance. The testing technique is a method of evaluating animals, such as performance and progeny tests. The performance test is a method of selecting candidate bulls based on their growth performance, whereas the process of using the carcass traits of their progenies to select candidate bulls is called progeny test (Chung et al., 2018). The performance and progeny test records comprise of pedigree, phenotypic and carcass information. These records are used for estimation of breeding values of individuals for proper selection. Based on the tests, 20 superior bulls are selected yearly and their semen is disseminated across the Hanwoo industry (Figure 2).
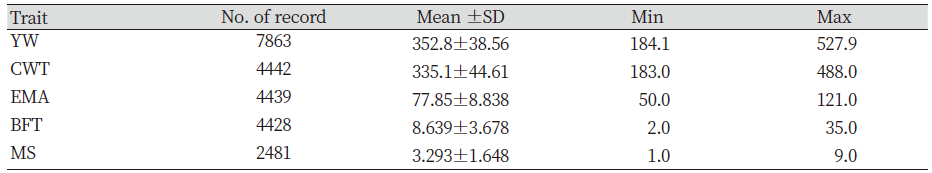
The HC possesses numerous economically important traits, but the emphasis was concentrated on the reproductive traits, growth performance and carcass traits. The improvement of reproductive traits focused on the high rate of pregnancy or conception rate and early puberty. The growth traits were focused on weaning weight and rapid growth rate. The carcass traits also received attention on higher dressing percent, lean meat, carcass weight (CWT), eye muscle area (EMA), marbling score (MS) and less backfat thickness (BFT) (Song, 1994).
To achieve the breeding objective, the Hanwoo production system was structured; comprised of the seed stock, cow-calf and feedlot sector. The Korean government, i.e., the operation of the Hanwoo breeding program, controls the activities in the seed stock sector. The cow-calf sector focuses on the multiplication of young calves through artificial insemination. The calves produced from the calf-cow are sold to the feedlot sector. Then, the feedlot sector raises the calves to slaughter age (Figure 3). With this system, a large number of animals are registered in the database and it encourages evaluation of genetic improvement as well as rate of genetic gain. The system further enables traceability on Hanwoo beef from the consumers’ endpoint to the Hanwoo farms (KAPE, 2016).
Breeding program for heifer replacements and cow selection
Over the years, attention has been given to bull selection and ways to improve the genetic makeup of the bulls in the beef industry. However, the beef cow and heifer replacements genetic improvement has not been a priority compared with bull selection. Ricky (2012) reported that there is a relentless decrease in beef cow numbers nationally, and it implies that a decrease in cow stock is associated with lesser feeder calves. The author further revealed that a lower feeder calf stock has put pressure on feedlot operators. Therefore, to sustain production, a significant and reasonable objective of each cow/calf operator should be to raise or sell 85 calves for every 100 cows consistently. This indicates that there would be a reduced number of culled cows because of greater reproductive efficiency present in the herd and as a result, increases herd income. It has been reported that reproductive performance is the main breeding objective for cow selection (Jonathan, 2016). Therefore, the improvement of reproductive performance could base on some of these factors: records keeping, such as percent calf crop, pregnancy rate, length of calving season, culling rates, calf morbidity and mortality, breeding efficiency of bulls, and performance and production information. More so, nutritional requirements, reproductive failure and abortions records should be incorporated. It is also important to select those individuals that have reliable calving records. To realize that a cow is reliable to calve, she should deliver her first calf early. It is obvious that breed, age and weight have a significant effect on puberty. In general, advantages exist for beef heifers that breed at 13-15 months and calve at 22-24 months; this implies that they draw the attention of the farmers by calving before the main herd begins to calve. In this way, they have additional time expected to rebreed with other matured cows. In all cases, heifers that bred at 14 months ought to have reached about 65-75% of their expected mature weight as a result, appropriate nutrition is essential.
Impact of genetic improvement on the performances of traits in Korean HC
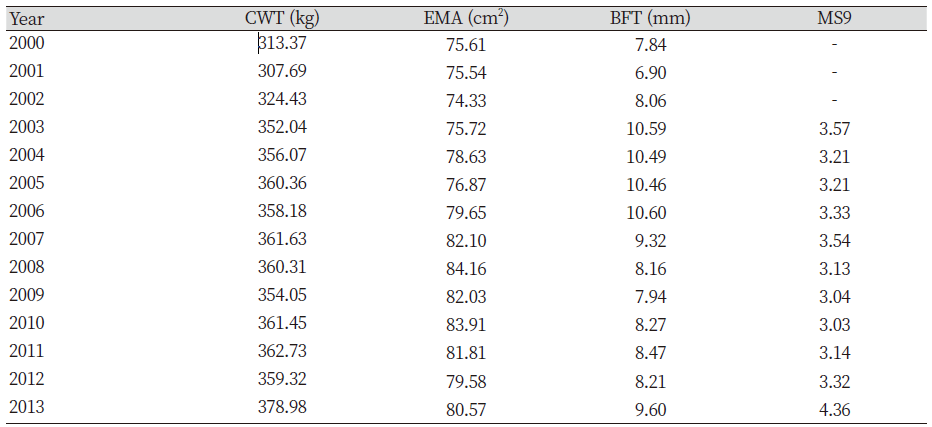
Genetic improvement has and continues to bring about considerable genetic progress to Hanwoo beef industry. This was possible through the breeding and production designs that provided the flow of genetic improvement where the genetic material from the nucleus herd is transferred to the feedlots. This design combines the visual appraisal, pedigree information, performance data and genetic merit value of the animals. Therefore, genetic improvement is a continuous process either towards the breeding objective or towards novel traits. For this reason, we compared the genetic trends and overall benefits of genetic improvement of previous and recent findings. The estimates of economically important traits for selected bulls were reviewed. The results show that the average body weight and daily gain at 12 months were inconsistent between the years (Table not presented). However, the findings also revealed that higher values were obtained from 1989 to 1992 (Song, 1994). Nwogwugwu et al. (2019) also reported a similar result in Korean Hanwoo cattle. On the other hand, Yoon et al. (2002) evaluated carcass records from 334 steer progeny of Hanwoo young bulls. The authors reported 301.1±34.3 kg, 74.8±8.7 cm2, 7.3±3.2 mm, and 3.04±1.60 in CWT, EMA, BFT and MS, respectively. The values reported by Yoon et al. (2002) were higher than those previously reported (Song, 1994). The Hanwoo Improvement Centre also has played a significant role in genetic progress over the years. The results from the Centre for weight of 18 months Hanwoo bull indicates the influence of genetic improvement. The findings indicated that the weight at 18 months in 1974 was 289.2 kg and increased to 552.8 kg in 2010, respectively. This scenario shows an increase of 263.6 kg for the past 36 years (Hanwoo Improvement Center, National Agricultural Cooperative Federation). Park et al. (2013) evaluated the yearling weight (YW) and carcass traits between 1998 and 2012. The authors reported 315.54 kg for YW in 1998 and the trait increased to 355.06 kg in 2011, resulting in about 40 kg of improvement over 13 years. Similarly, a report from the Hanwoo testing program also showed improvement in the studied traits. Further findings reported increased values of marketed weight, CWT, EMA and MS in 10 years by 109 kg, 74 kg, 9.1 cm2 and 1.8 (KAPE, 2012). Park et al. (2013) also compared average values of numerous traits with previous studies (Table 1). The results of the YW and carcass traits showed improvement compared with previous reports. Nwogwugwu et al. (2019) reported increased values of carcass traits from 2000 to 2013, which shows the overall improvements in most of the traits (Table 2). Note that the results of these estimates depend on the source of datasets because some of the records may come from HIC, private farms or abattoirs.
Evaluation of genetic improvement
Pedigree and performance records
Genetic improvement programs for cattle and other livestock species were based on pedigree and performance records. Pedigrees are essential instruments in the livestock breeding because they provide information on the ancestors and knowledge for predicting offspring performance. This implies that a pedigree record comprises the performance records of individuals and their progeny; as a result, it reveals the traits of economic importance for each domesticated animal species. For example, the meat and milk traits in cattle, sheep and goats (NPLC, 2019). Carmen (2007) reported that pedigree was earlier introduced in cattle breeding and other domestic animals. Henceforth, it becomes the main breeding tool in the livestock sector. Previously, pedigree and performance records have been used to evaluate the genetic improvement of HC. This implies that superior bulls are selected via their estimated breeding values (Lee et al., 2014).
Estimation of breeding values
Various methods have been used for estimating genetic breeding values of individuals. The EBVs are usually used for selecting superior individuals. The BLUP (Best Linear Unbiased Prediction) is one of the methods used to estimate breeding values, and it uses an animal model (Henderson, 1975; Xu-Qing Liu et al., 2008). The BLUP procedure makes use of the performance records of related individuals within the same or different herds to estimate breeding values and environmental effects at the same time. With quite numbers of related animals in different groups and years, the BLUP EBVs could be compared across groups and years. This suggests that the BLUP would also provide higher accuracy of EBVs compared to some methods of breeding value estimation. The BLUP further takes into consideration of fixed and random effects. Applying BLUP is a potential method to evaluate the EBVs of carcass traits for genetic improvement before selecting superior bulls annually (Figure 2).
Factors influencing genetic improvement
The success of selection is usually measured by the rate of genetic improvement. For this reason, it is essential to identify a few factors that can influence genetic change through selection. Reports have identified numerous factors, such as mating system, population size, methods of prediction, pedigree errors, selection accuracy, selection intensity and the generation interval (Van der Werf, 2015; Nwogwugwu et al., 2019). In this study, four factors were explained: selection accuracy, selection intensity, pedigree errors and generation interval.
Selection accuracy is associated with the prediction accuracy of EBVs. The more accurate the EBVs, the more possible that the animals chosen to be the parents would be the best (AIGR, 2016). The accuracy of selection also depends on the heritability of the trait. That is, the higher the heritability, the better the prediction accuracy. To increase the selection accuracy, we need good information about the candidates for selection because the only existing information is the phenotypic records, which is the strength of the relationship between phenotypic values and breeding values. It has been reported that other factors such as environmental effects and contemporary groups should put into consideration (Henderson, 1975; Xu-Qing Liu et al., 2008; Nwogwugwu et al., 2019).
Selection intensity determines which individuals are to be selected from a population. This factor creates pressure on the breeders, but if the selection criteria are practically precise, then the selected parents should be genetically better than average. During this change, the rate of genetic improvement should be fast with superior animals in the next generation (AIGR, 2016).
Pedigree errors (PEs) could affect the accuracy of prediction as well as the rate of genetic improvement. Harder et al. (2005) earlier reported two types of pedigree errors, which could affect the EBV and genetic gain in dairy cattle. Several authors have also reported the effect of pedigree errors (missing information about the parents) and wrong parentage on the accuracy of EBV and genetic parameters (Christensen et al., 1982; Gelderman et al., 1986; Israel and Weller, 2000). For example, reduction in the value of a parent transmitting ability (PTA) for a cow and her relatives; reduced EBV, heritability (h2), and genetic gain for meat and milk traits in cattle. More recently, Nwogwugwu et al. (2019) reported reduced accuracy of EBV, h2, and genetic gain for carcass traits in Korean HC. In contrast, the use of genomic information could improve the accuracy of genomic estimated breeding values when errors occurred in the pedigree dataset (Unpublished article). It has been reported that genomic evaluation methods improved the prediction accuracy in genotyped and non-genotyped individuals (Meuwissen et al., 2016).
Generation interval (GI) measures the time required to change one generation to another. Shorter GI encourages quick rate of genetic change, although, environmental and biological factors might influence the GI, such as management, species and reproductive rate. The GI in beef cattle is longer than in small ruminants; as a result, the gestation period, the weaning period, and the age at puberty is longer. For these reasons, genomic selection could be a tool to reduce the generation interval. The use of GS has revealed a remarkable change in GI in the United States Holstein cattle (García-Ruiz et al., 2016). The authors reported a reduction in GI from almost 7.5 years to less than 2.5 years in the sires of bulls’ pathway. The findings further reveal a decline from 7 years to less than 5 years in the sires of cow’s pathway and such a scenario increased the rate of genetic gain.
Implementation of genomic selection in Korean HC breeding
The completion of cattle genome and other livestock has altered the shape of breeding programs based on the genomic breeding value by applying single nucleotide polymorphism (SNP) information. Previous studies have reported improved genetic gain, enhanced prediction accuracy of GEBVs and reduction in generation interval (Hayes et al., 2009; VanRaden et al., 2009; Harris and Johnson, 2010; Su et al., 2012; Meuwissen et al., 2016). Calus (2010) reported that the efficiency of GS in livestock depends on the prediction accuracy of GEBVs. Mrode (2005) earlier reported the advantages of using de-regressed breeding values over the use of actual phenotypes for developing training populations (TP) in dairy cattle. He revealed that the method significantly produces phenotypes with high heritability that is ideal for accurate prediction of GEBVs. Compared with Holstein-Friesian dairy breeding, the Korean HC breeding scheme appears to have similar structures susceptible to GS, especially considering the influence of the proven bulls and commercial breeding. This scenario creates a link to transfer genetic materials from elite bulls to other sectors that can produce accurate GEBVs. In addition, the extensive use of AI in Hanwoo breeding would provide an ideal situation and cost-effective in the TP size and easy validation of genomic tools through existing progeny testing. This process will further provide speedy distribution of genetic merit to the feedlot sector (Figures 2 & 3).
Potential for implementation of GS in Korean HC
Several authors have reported the benefits and potential for implementation of GS in dairy cattle. In the Hanwoo breeding scheme, few existing structures would encourage implementation and evaluation of GS. First, the establishment of genetic composition exists in Hanwoo breeding scheme (Figure 2). It is also important to note from Figure 2 that 20 proven bulls are selected per year, and this technique does not create a diverse range of pure breeds in the seed stock sector. With a well-organized and known breeding structure, the majority of the animals are registered and animals’ records are traceable from the consumer’s table to individual farms. It has also indicated that the breeding method encourages wide use of AI, which is a driving force behind the implementation of GS. This implies that an ideal state of TP with large numbers of AI sires will result in a higher accuracy of GEBV in the latter sectors. Therefore, higher accuracy of GEBV will encourage a reduction in GI leading to quicker genetic gain. More so, the availability of performance records, pedigree and progeny test information is an added advantage in the sector (Lee et al., 2014). The existence of a national cattle database in Korea permits the flexibility and accessibility of animals’ records (KAPE, 2016). These few points mentioned above will encourage the implementation of GS in Hanwoo breeding scheme.
Prediction accuracy of GEBV through genomic selection
Genomic selection increases the prediction accuracy of GEBVs in livestock species. Several authors have reported numerous studies based on GS. The two-step genomic best linear unbiased prediction (tsGBLUP) has been used to evaluate the prediction accuracy of GEBVs in dairy cattle. This method combines direct genomic value (DGV) and EBV from phenotypic records to estimate the prediction accuracy of GEBVs. The combination could also help genomic markers capture any QTL effect or polygenic effect through EBVs (Hayes et al., 2009; VanRaden et al., 2009; Pryce et al., 2014). The single step best linear unbiased prediction (SSGBLUP) predicts how non-genotyped individuals can benefit from genomic information. The pedigree record and marker (SNPs) relationship matrices are combined in SSGBLUP, permitting the blending of genotyped and non-genotyped individuals in the genetic evaluation (Legarra et al., 2009). However, Onogi et al. (2014) and Cardoso et al. (2015) reported that the use of SSGBLUP for genomic evaluations is still emerging in beef cattle. Numerous studies have revealed that genomic evaluations produce higher prediction accuracy than the traditional BLUP in sheep (Daetwyler et al., 2012) and beef cattle (Gowane et al., 2018). Based on these authors, GS improves the prediction accuracy as well as genetic gain of the traits. In Korean HC, the reports on genomic evaluations are limited. However, a comparison test has been performed between two methods of genetic evaluations (TBLUP and GBLUP) on the prediction accuracy of EBVs or GBEVs in Hanwoo cattle. The results showed that the prediction accuracy of GEBV was higher than the accuracy of EBV (Lee et al., 2014). Reports also suggest that GS could be used to improve low heritability traits and those traits that are difficult or expensive to measure (Meuwissen et al., 2016). Mehrban et al. (2017) reported increased accuracy of DGVs in carcass traits using different evaluation methods in Hanwoo beef cattle. However, the prediction accuracy of GEBVs can be influenced by several factors, such as the heritability of the trait (Meuwissen et al., 2001), the extent of linkage disequilibrium (Brito et al., 2011), the TP size (Calus, 2010), the marker density (Solberg et al., 2008), the effective population size (Hayes et al. 2009) and the methods of prediction (Gao et al., 2012; Lourencol et al., 2014; Mehrban et al., 2017).
Sometimes, it might be difficult to understand the manners those factors influence the prediction accuracy of GEBVs. However, the used of simulation study has helped to reveal different scenarios. A simulation study allows the testing of several theories, permitting an unravelling of the complex evolutionary patterns that are otherwise difficult to comprehend (Rogers et al., 2007; Marchani et al., 2007). Simulation studies in beef cattle and other livestock have provided information on their potential for genomic evaluation. Numerous studies have reported higher prediction accuracy of GEBV compared with the accuracy of EBV in simulated beef cattle (Piccoli et al., 2018) and American mink (Karimi et al., 2019). Recent study by Nwogwugwu et al. (Unpublished) further revealed that genomic selection would encourage improvement of low heritable traits. Based on these findings, genomic tools hold the potential to increase genetic merit in Hanwoo cattle breeding program.
Prospect for genomic selection
The future seems positive and brighter with GS in some livestock industries; however, considerable attention is required toward the economic viability. Selection for new traits are essential, but they may be difficult or expensive to measure. Traits such as feed efficiency (FE), disease resistance, meat quality and greenhouse gas (GHG) emission are difficult to select (Todd, 2013). The author further explained several sectors that could assist in selecting of those traits, such as the commercial breeders, finishers and supermarkets or consumers. These sectors might be a driving force where detailed information about the animals and their products could be used for genomic evaluation. As pointed out by Berry and Crowley (2011) FE and its derivative, such as residual feed intake (RFI) is one of the areas of interest in animal science research. It has been reported that those traits have moderate heritability, expensive to measure and have significant economic influence (Herd et al., 2002; Van der Werf, 2009). For GHG, it has been revealed that animal breeding might be a way out for selecting those traits that have genetic correlation (Alford et al., 2006; Hegarty et al., 2007). Nowadays, meat quality seems to be an important factor for consumers. The application of GS indicates impressive results for selection of meat quality trait in beef cattle (Van Eenenneem et al., 2011; Pimentel and Konig, 2012). The authors suggested that GS could significantly increase the genetic gain. Respect to disease resistance traits, the application of GS would be a huge success. Studies suggested that GS is a possible technique for selecting disease resistance traits in fish and poultry (Nirea et al., 2012; Fulton, 2012). In this regard, Korean HC breeding and production systems could explore GS to improve most of the traits.
Conclusion
Genetic improvement requires proper selection of superior individuals through estimation of breeding values. To buttress genetic improvement, it is important to define the breeding objective, production and breeding systems. Evidence from our study indicates that there is a need for proper recording to enable evaluation of genetic progress. The results of the previous and recent studies for carcass traits indicate the overall genetic improvement. In domestic livestock, such as Korean Hanwoo cattle, the application of GS could be a possible way to improve meat quality, disease resistance, and feed efficiency traits.