Mechanisms of HDR and NHEJ pathway
HDR pathway is one of the essential mechanisms to maintain genome integrity because of its high fidelity of DNA repair. DNA double strand breaks (DSBs) repair by HDR pathway is mainly conducted in mitotic cells of the late S or G2 phase of cell cycle, because HDR pathway repairs DSB by using sister chromatid as template (). Because the sequence of sister chromatid is identical to damaged DNA strand, HDR pathway can repair DSB accurately and maintain intact genetic information without error. When DSB occurs, the exonuclease recognizes DSB and makes 3’ single-stranded DNA (ssDNA) overhangs by trimming process. After ssDNA formation, RAD51 recombinase (Rad51) binds to ssDNA and promote DNA strand invasion into the homologous sister chromatid. After alignment between invaded DNA strand and sister chromatid, the polymerase initiates DNA synthesis from 3’-OH end of ssDNA using sister chromatid as template. Subsequently, the ligase joins the nick of the each strand (). This HDR pathway can be applied to insert the foreign genes of interest (GOI) into the target sites. When donor plasmid containing GOI contains homology arms of target site, the HDR pathway recognizes the homology arms of donor plasmid as template and insert GOI precisely into the target site.
On the other hands, NHEJ is predominant DSB repair pathway which does not require homologous sister chromatid (). When DSB occurs, the NHEJ machineries detect the damaged site, modify and rejoin broken end of each DNA strands. Firstly, the Ku70-Ku80 heterodimer protein binds to broken ends and forms synapsis, which allows alignment of the two broken ends and subsequent repairing process (). The formation of synapsis facilitates recruitment of enzymes such as DNA nuclease, polymerase, and ligase. The DNA nuclease and polymerase conduct nucleotide excision and addition at each of the two strands to make micro-homology. After the micro-homology is formed, the two strands are aligned and the gaps are filled by polymerase. After filling the gaps, the ligase joins each nick of the two strands (). During this process, deletions or insertions of a few nucleotides occur in the damaged site, results an insertion-deletion (indel) mutation on the site. Because of this error-prone manner, NHEJ mediated DNA repair generates frame shift mutation of gene and disrupts of gene function.
Genome modification based on DNA repair pathway
Based on the mechanism of DNA repair pathway, investigators have tried to apply it for genome modification in living cells. Lin et al. firstly identified that when the foreign DNA introduced into the mouse fibroblast cells by calcium phosphate method, introduced exogenous DNA was inserted into the genome by HDR pathway (). Furthermore, the mutant form of neomycin resistant gene was corrected by HDR mediated gene targeting (), and the mutant form of Hprt gene, which encodes protein necessary to generation of purine nucleotides, was replaced by HDR mediated gene targeting and the function of the gene was restored (). Subsequently, Thomas and Capecchi induced mutation in Hprt gene of mouse embryonic stem (ES) cells and successfully produced germline chimera by injecting the gene-targeted ES cells into the blastocyst (). This result demonstrated that gene targeted organism can be generated by HDR mediated gene targeting in ES cells, although the efficiency of HDR mediated gene targeting was extremely low (one genome-modified cell per 104~108 cells) (; ). After that, Rouet et al. observed that inducing of DSB accelerated DNA repair process at the damaged site and facilitated HDR mediated genome editing () and this report subsequently promoted development of programmable genome editing technologies for efficient genome modification.
Programmable genome editing tools for genome modification
Programmable genome editing (PGE) tools such as zinc finger nucleases (ZFN), transcription activator-like endonucleases (TALENs) and clustered regularly interspaced short palindromic repeat (CRISPR) and CRISPR-associated protein 9 (Cas9) system (CRISPR/Cas9) induce DSB at target site efficiently and enhance HDR or NHEJ mediated genome modification. ZFN consists of DNA binding zinc finger motif and FokI endonuclease enzyme. Each zinc finger motifs can bind three nucleotides, therefore it can be modulated to bind target DNA sequence by changing combinations of amino acid sequences of zinc finger motifs (). ZFN has been widely used to produce genome modified organisms in various species (). However, the limitation of ZFN is frequent lack of DNA targeting activity and high frequency of off-target effects (; ). TALEN consists of TAL effector derived from Xanthomonas spp. and FokI endonuclease. TAL effector consists of 33-34 amino acids with repeat variable di-residue (RVD) in the 12th to 13th position of amino acids (). Unlike to zinc finger domain, each TAL effector can recognize one nucleotide, so it is more flexible to design target specific TALENs than ZFN (). Based on the versatility, TALENs has also been widely applied to produce genome modified organism of various species (). However, the difficulty of protein engineering and high cost of designing TALENs prevented broad application of TALENs in genome editing (; ).
Currently, the CRISPR/Cas9 system has become highly versatile and convenient tool to accomplish targeted genome modification. Originally, CRISPR/Cas9 system is derived from bacterial immune system which binds and disrupts invaded viral RNA segments (). When CRISPR RNA (crRNA) and trans-acting CRISPR RNA (tracrRNA) bind to target nucleotide sequences, Cas9 recognizes and cleaves targeted nucleotide sequences (). These two crRNA and tracrRNA can be engineered as one single guide RNA (sgRNA) (). As described above, CRISPR/Cas9 system only needs sgRNA that binds to target site and Cas9 protein which recognizes guide RNA and cleaves DNA double strands for inducing DSBs in targeted loci. Because of convenience, nowadays, the CRISPR/Cas9 system has become the most widely used tool in genome modification (). Using this powerful genome editing tool, it has become possible to modify various functional genes of genome to produce novel organisms possessing functional traits. Concurrently, many researches have been conducted to enhance the efficiency of targeted genome modification to facilitate wide application of genome editing technology.
Applications of CRISPR/Cas9 for genome editing
1) HDR mediated genome editing
Using CRISPR/Cas9, various genome-edited model organisms such as worm (Caenorhabditis elegans), fly (Drosophila melanogaster), zebrafish, mouse, chicken, and pig have been developed. Since CRISPR/Cas9 mediated DSB promotes HDR pathway in living organisms, investigators have applied the genome editing tool to enhance HDR mediated genome modification. In C.elegans, Chen et al. firstly inserted hygromycin resistance gene in the C. elegans genome by HDR mediated targeted gene insertion (). In mice, HDR mediated gene targeting of Tet1 and Tet2 genes which regulate DNA methylation and gene expression has been accomplished by injecting Cas9 mRNA, sgRNA and ssDNA oligos into one cell embryo (). Also, mice carrying a fluorescent reporter construct in the pluripotency related genes such as Nanog, Sox2 and Oct4 was produced by HDR mediated gene targeting (). In addition, to produce mouse model which can expresses foreign genes stably, the Rosa26 locus targeted mouse model which can expresses foreign genes stably was produced via CRISPR/Cas9 system (). In chicken, CRISPR/Cas9 mediated targeted genome modification was applied to produce genome modified bird which possesses human heavy chain constant region for bio-medical application (). Also, the chicken which accumulates human IFN-β in the egg white has been generated by CRISPR/Cas9 mediated HDR (). In pigs, HDR mediated targeted gene insertion of uncoupling protein 1 (UCP1) improved cold adaptation of pig and decreased fat deposition in the muscle (). Taken together, HDR mediated genome modification has been widely applied to introduce novel phenotypes into organisms.
Although HDR can be applied to introduce valuable traits into organisms, HDR mediated genome editing is inefficient because NHEJ is predominant repair pathway throughout the cell cycle, while HDR occurs during the late S and G2 phases of the cell cycle (). To improve efficiency of HDR mediated genome editing, there have been various attempts to overcome the low efficiency of HDR for the past few years.
It has been reported that the efficiency of HDR can be improved by inhibiting NHEJ pathway (; ). Recently, Chu et al. demonstrated that suppression of DNA ligase IV, which is critical enzyme for NHEJ, increased efficiency of HDR by 4~5-fold. They also discovered that expression of adenovirus proteins such as E1B55K and E4orf6, which promote degradation of DNA ligase IV, improve the efficiency of HDR up to 8-fold (). Furthermore, it was also reported that suppression of DNA ligase IV using small molecule, Scr7, can increase the efficiency of HDR by up to 19-fold ().
Modulating cell cycle is another strategy to increase the efficiency of HDR. Synchronizing the cells at G1, S and M phase by treatment of chemical inhibitors (aphidicolin, hydroxyurea, lovastatin, mimosine, nocodazole, and thymidine) increased the rates of HDR up to 38% (). Another research group developed human Cas9 (hCas9)-human Geminin (hGem) by fusing hCas9 to the N-terminal region of hGem, which is the direct target of the E3 ubiquitin ligase complex APC/Cdh1. Since the activity of the APC/Cdh1 E3 is high in the late M and G1 phases, the genome modification by hCas9-hGem is restricted to the S, G2, and M phases, resulting in increased efficiency of HDR up to 87% ().
To enhance the efficiency of HDR mediated genome editing, many efforts have been focused on the identification of the specific chemicals or small molecules which can improve the rate of HDR. By a high-throughput screening of 4,000 small molecules, the two molecules L755507 and Brefeldin A have been identified to increase the HDR efficiency (). Recently, it was reported that RS-1 which can stimulate human homologous recombination protein RAD51 also increases efficiency of HDR mediated knock-in by 2~5-fold in rabbit (). More recently, the treatment of NU7441 which can suppress NHEJ pathway improved HDR mediated gene modification up to 13.4-fold ().
For precise modification of the target loci by introduction of exogenous DNA, it is necessary to transfect exogenous donor DNA to the cell. The efficiency of HDR can be improved by optimal design of donor DNA. The single-stranded oligodeoxynucleotides (ssODNs) were identified as efficient donor DNAs for ZFN-mediated genome editing (; ). Since then, ssODNs are utilized in CRISPR/Cas9-mediated genome editing (; ; ). Meanwhile, Aird et al. fused ssODN with the Cas9-guide RNA ribonucleoprotein (RNP) complex. This way, they enhanced HDR efficiency up to 30-fold (). Also, Zhang et al. showed that when HDR donor plasmid is double cut by CRISPR/Cas9 by inserting sgRNA sequences, the efficiency of HDR improved by 2-5 folds than using circular donor plasmid as HDR donor. Furthermore, the 97-100 % donor insertion events were mediated by HDR when donor plasmid contains 600bp of homology arms ().
2) NHEJ mediated genome editing
It was showed that NHEJ mediated targeted gene insertion is accomplished when chromosome and donor plasmid has ZFN or TALEN target site concurrently (; ). It means that if chromosome and donor plasmid has sgRNA target site, Cas9 will cleave chromosome and donor plasmid concurrently and donor plasmid will be inserted into sgRNA target site of chromosome. NHEJ mediated targeted gene insertion using CRISPR/Cas9 system was firstly reported in the zebrafish (). The investigators used eGFP transgenic zebrafish and they designed sgRNA specific to eGFP transgene and donor plasmid which includes transcriptional trans-activator Gal4 (KalTA4) and sgRNA recognition site. When they co-injected sgRNA/Cas9 mRNA and donor plasmid into one-cell stage zebrafish embryos, they observed targeted insertion of donor plasmid into eGFP transgene locus and loss of eGFP expression. Also they observed that the sequences of the 5’ and 3’ junction site showed the indel mutation. Similarly, Kimura et al. conducted targeted gene insertion in zebrafish by injecting donor plasmid containing heat shock protein promoter, reporter gene, and sgRNA target site into the one cell stage embryo with Cas9 mRNA (). The authors observed that the efficiency of obtaining transgenic founders is over 25%. These results demonstrated that CRISPR/Cas9 efficiently mediates integration of donor plasmid by NHEJ pathway. Remarkably, He et al. showed that the efficiency of reporter gene integration of NHEJ mediated targeted gene insertion is significantly higher than that of the HDR mediated targeted gene insertion in various human cells including human ES cells (). Subsequently, Suzuki et al. integrated donor plasmid into the genome of non-dividing cells such as neurons using NHEJ mediated targeted gene insertion. They successfully conducted in vivo genome editing by injection of adeno-associated virus (AAV) containing donor plasmid, Cas9, and sgRNA expressing vector in brain (). These results showed that foreign genes can be efficiently integrated into the genome of the dividing and non-dividing cells by NHEJ mediated targeted gene insertion and this strategy can be applied to various species.
Recently, the genome modified chicken based on NHEJ mediated targeted gene insertion using CRISPR/Cas9 system has been reported (). In this report, the investigators established a novel genome modified chicken line which contains GFP expressing gene cassette in the Z chromosome to develop avian sexing model. They constructed donor plasmid which has two same sgRNA recognition sites at each end of the gene cassette. After transfection into chicken primordial germ cells (PGCs) with Cas9 expression vector and donor plasmid, they detected GFP expression in PGCs and validated targeted integration of donor plasmid into Z chromosome. After that, they transplanted genome edited PGCs into the embryo and subsequently produced eight donor PGC derived progenies and five of them (62.5%) were Z chromosome targeted gene insertion via NHEJ pathway.
Above results showed that the efficiency of gene insertion via NHEJ is higher than that of HDR, although there are some indel mutations at junction site.
Conclusion and Future Perspectives
As described above, efficient targeted genome modification methods have been developed for the last few decades. Thanks to effort of pioneers in biotechnology, now the targeted genome modification technology is expected to develop various industrial fields as well as basic science area. In agriculture, targeted genome modification can confer resistance to environmental threat such as temperature and pathogens, and improve growth performance of crops. In animal industry, targeted genome modification can generate animals with higher economical traits such as disease resistance, cold or thermal resistance, enhanced meat production, enhanced feed conversion rate, and meat with enhanced nutritional value. Animal bioreactors, which can produce recombinant bio-therapeutics with higher efficacy economically, can be produced by targeted insertion of functional genes in the locus of egg white protein genes or milk protein synthetic genes (; ). Also, targeted genome modification now has been applied to restoration of extinct species such as mammoths (). For extermination of vermin, scientists produced genome modified mosquitoes which have defect in their reproductive function (). In the future, genome modification technologies will be consistently developed and will improve human welfare and scientific knowledge by bringing innovations in various fields of industry and basic science.
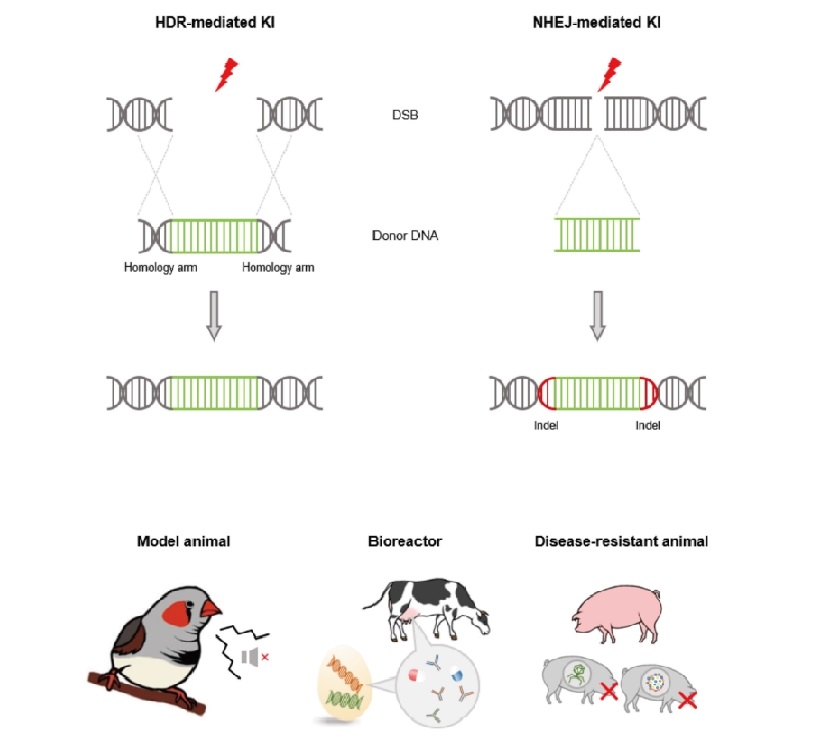
Figure 1. Targeted genome modification mediated by DNA repair pathways and its application. The two major DNA repair pathways (HDR and NHEJ) are initiated when DSBs occurs by programmable genome editing tools such as CRISPR/Cas9. When donor plasmid exists, DNA repair pathways mediate targeted genome modification. Based on the genome modification technology, various organism can be generated such as model animal, animal bioreactor, disease resistant livestock and other novel organisms which have valuable traits.


