Abstract
The objective of the study was to identify signatures of divergent selection associated with production and adaptive traits in Ethiopian and Hanwoo cattle by using a high-density and informative SNPs (80K) specifically developed from Bos indicus breeds. The most highly differentiated regions representing the top 1% windows were used to define genomic regions under divergent selection. Based on population genetic differentiation ( FST) analysis, a total of 334 genes were identified in the comparison of Ethiopian cattle with Korean cattle (Hanwoo). The genes are associated with thermotolerance (DNAJB5/Hsp40), pigmentation (ALX3, DCTN2, and MYO1A), reproduction/fertility (INHBC, UBE2D3, ID3, PAIP2, and PSPC1), and body stature and production traits (MBP2, HERC3, and GJB3 and POLR2A) and carcass quality (TFEC, MYOT, and PARPA ). Functional enrichment analysis further showed that the candidate genes were associated with pathways relevant for physiological adaptation, including vasopressin-regulated water reabsorption, Rap1 signaling, PIK3 signaling, and melanogenesis to extreme environments. By comparing cattle breeds with distinct evolutionary and breeding legacies, we spotted candidate genes under selection and pathways which may contribute to further understanding of the consequence of natural and/or artificial selection in molding the phenotypic diversity of modern cattle breeds.
Figures & Tables
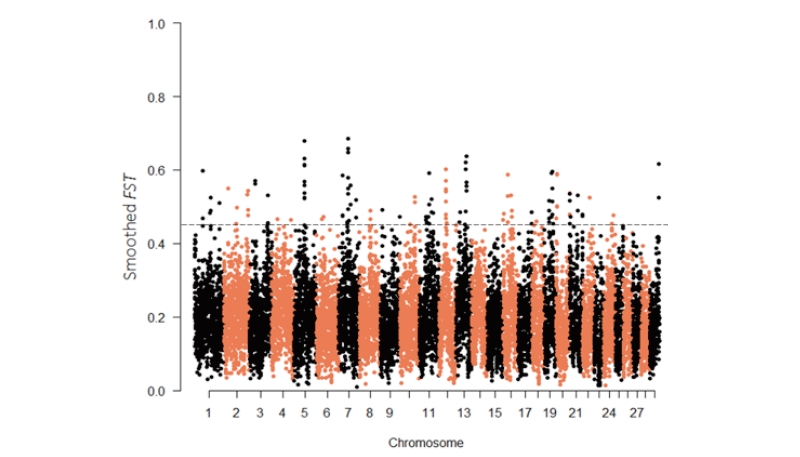
Figure 1. Manhattan plots of genome-wide smoothed F values. The dotted gray line indicates the top 1% F values of the genome-wide empirical distribution


