Abstract
This study was conducted to compare the accuracies of estimated breeding value (EBV), genomic estimated breeding value (GEBV) and single-step genomic estimated breeding value (ssGEBV) for economic traits of 935 Hanwoo cows. The economic traits considered in this study were carcass weight (CW), eye muscle area (EMA), backfat thickness (BFT) and marbling score (MS). The EBV analysis was performed using the best linear unbiased prediction (BLUP) method by constructing a numerator relationship matrix (NRM) with the pedigree information of 935 cows and 9,849 Hanwoo steers with phenotype data. The GEBV analysis was performed using genomic best linear unbiased prediction (GBLUP) method by constructing a genomic relationship matrix (GRM) with SNP 50K information of 935 cows, and phenotypic and genomic data of Hanwoo steer used in BLUP analysis as reference population. The ssGEBV analysis was performed with single-step genomic BLUP (ssGBLUP) based on the relationship matrix H, which is constructed from the numerator relationship matrix (A) augmented by the genomic relationship matrix (G). As the results, the differences in accuracies of GEBV and EBV for CW, EMA, BFT and MS traits for 935 cows showed that the accuracies of GEBV were increased by 47.17%, 42.56%, 44.13% and 51.17%, respectively. The accuracies of ssGEBV compared to EBV for CW, EMA, BFT and MS were increased by 47.64%, 43.02%, 44.60% and 51.41%, respectively. When ssGEBV was compared with GEBV for CW, EMA, BFT and MS, there were increased by 0.32%, 0.33%, 0.33% and 0.16%, respectively. As conclusion, this study suggests that GBLUP and ssGBLUP methods for Hanwoo cows are able to improve the accuracy of the estimated breeding value using genomic information. It could to help the management of cows and the establishment of a breeding system in the Hanwoo farms.
Figures & Tables
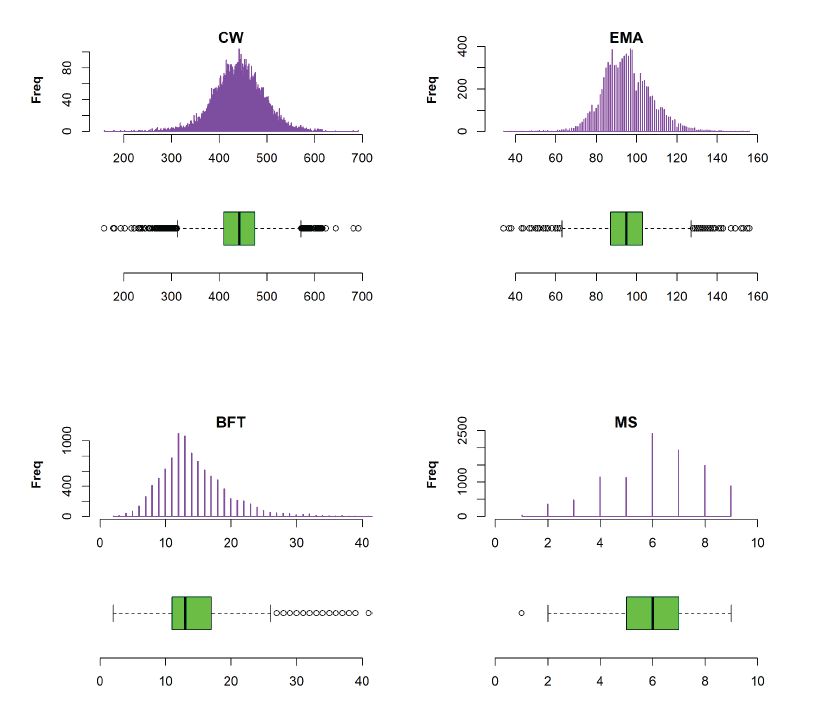
Fig. 1. Distribution and quantile-quantile plot (Q-Q plot) for carcass traits in 9,849 steers.


