Abstract
Based on coat color phenotypes, Korean native cattle are categorized into four breeds/strains (Brown Hanwoo, Black Hanwoo, and Brindle Hanwoo) and each strain has experienced different demographic events. In this study, to decipher the patterns of genetic diversity and identify signatures of selection, we genotyped 284 animals representing 3 Korean cattle breeds using the 600K Affymetrix chip. The lowest within-breed genetic diversity was found for Brindle Hanwoo (Chikso) and Black Hanwoo (Heugu) cattle, probably a result of a reduction in effective population size. Our principal component and phylogenetic analyses revealed a marked clustering of the three cattle breeds according to their phenotypic /coat color basic discriminations. The mean estimates of r2 were 0.30, 0.29 and 0.29 in the Chikso, Heugu, and Hanwoo cattle, respectively. The Chikso and Heugu breeds reliably showed smaller Ne across generations. Our signatures of divergence selection analyses identified functionally important genes. Notably, some of the candidate genes have previously been known to affect growth, body size and stature (GLI3, XKR4, CDK6, DIAPH3, ELF1, FTO), meat quality (XKR4, DECR1, FTO, DIAPH3), fatty acid metabolism and composition (ELF1, PPARD, MTTP), and feed efficiency (XKR4, NR1I2). Gene Ontology(GO) analysis showed that that the candidate genes were associated with functional terms relevant to meat quality traits (fatty acid beta-oxidation, adipose tissue development) and pigmentation. The genetic diversity and structure of the three Korean cattle breeds can be explained by demographic events, remarkably genetic drift and selective breeding. It can be suggested that genes with known effect on growth and meat quality traits are potential candidates for further validation and inclusion in the beef selection program.
Figures & Tables
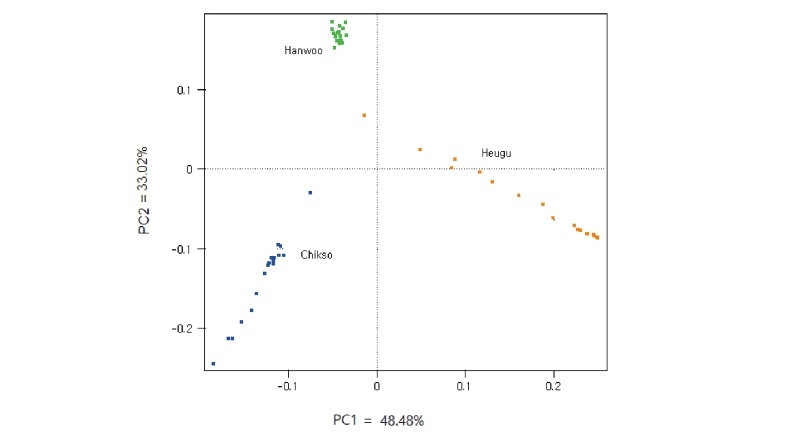
Figure 1. Individual animals clustering based on principal component analysis (PCA)


