Abstract
Genomic selection has forever changed the field of animal breeding, and this technology has had a remarkable impact on dairy cattle improvement programs, where long generation intervals and sex-limited traits add complexity and expense to conventional pedigree-based selection schemes. Tens of thousands of single nucleotide polymorphism (SNP) markers dispersed across the genome can be used to track the inheritance of chromosomal segments from one generation to the next, and these markers tend to be inherited with nearby and typically unknown quantitative trait loci (QTL) that affect the phenotypes for traits of economic importance on dairy farms. More importantly, knowledge of the relationships between SNP markers and nearby QTL in older reference population animals with both genotypic and phenotypic data can be used to predict the genomic estimated breeding values (EBV) of young selection candidates for which only genomic test results are available. Development of statistical methods for estimation of SNP effects and prediction of genomic EBV, as well as methods for imputation of inexpensive low-density SNP genotypes to higher density, has been rapid. This technology has been adopted quickly in North America, where approximately two million dairy cattle have been genomic tested over the past eight years. The rate of genetic progress for most traits has doubled, due to much shorter generation intervals for the sires of males, sires of females, and dams of males selection pathways, in addition to greater accuracy of decisions involving female selection candidates. The next frontier in genomic selection is development of routine predictions for novel traits that are too difficult and expensive to measure in traditional milk-recording programs, such as feed efficiency, calfhood pneumonia, and early postpartum health. In the long-term, genome-guided decision tools offer promise as regards the management of animal health, performance, and welfare, but significant research is needed to make these opportunities, which are the bovine equivalent of personalized medicine, technically feasible and cost-effective on commercial dairy farms.
Acknowledgements
This work was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ012078)” Rural Development Administration, Republic of Korea.
Figures & Tables
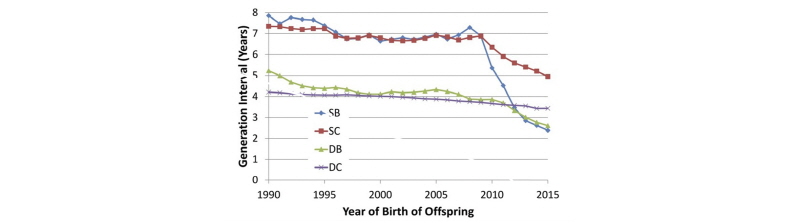
Generation intervals for the four paths of selection in U.S. Holsteins: sires of bulls (SB), sires of cows (SC), dams of bulls (DB), and dams of cows (DC), from García-Ruiz et al. (2016).
