Abstract
This study was conducted with the Hanwoo SNP chip (50k_v1) analysis to investigate genotypes of SNPs of Hanwoo (Korean native cattle) in Jeollabuk-do and extent of linkage disequilibrium (LD) among SNP markers. Genomic DNA for genotyping was extracted from the hAir root samples of Hanwoo cows (n=1,523). A total of 52,195 SNPs were obtAined and 11,056 unnecessary SNPs were removed through the QC process, and a total of 41,139 SNPs were used for LD analysis. The total length of the SNP markers used in the analysis was 2,500 Mb, the mean of minor allele frequency was 0.27 and the average interval distances of adjacent SNP markers by each chromosome was 0.0463 to 0.0725. The results of this study which confirmed by using Hanwoo in Jeollabuk-do, showed a pattern similar to previously reported studies for Hanwoo. The r 2 value was 0.234 for the SNP distance between 0 and 50 Kb and was 0.068 for distance between 150 and 200 Kb. Therefore this study could be utilized as baseline data using genetic improvement for Hanwoo industry.
Figures & Tables
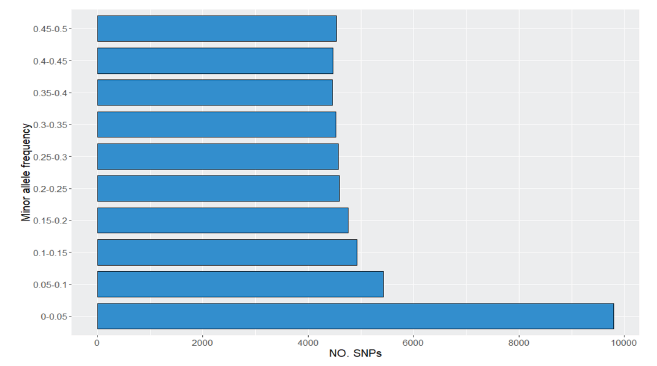
Fig. 1. Minor Allele Frequency of SNPs


