Abstract
Olive flounder (Paralichthys olivaceus) is one of the most produced fish, accounting for 46.2% (37,240 metric tons) of total fish farming yield in Korea. Flounder is vulnerable to viral hemorrhagic septicemia (VHS) which makes fish farmers struggle. Viral Hemorrhagic Septicemia Virus (VHSV) is an RNA virus sustaining its infectivity, especially at low temperatures (8-15°C). In order to minimize the damage from VHSV, several studies were conducted. However, the genetic factor of resistance to VHSV is not well known to date. Quantitative trait loci (QTL) analysis is an efficient analysis in aspects of detecting loci that influence quantitative traits. We conducted a quantitative trait locus (QTL) analysis by constructing a genetic linkage group. 23 linkage groups were constructed and it’s reliable according to the previous study that suggested flounder has 2n=48 chromosome. Bivariate phenotypes (dead/alive) were adapted and 5 pseudo markers were identified as QTL. A total of 7 markers were nearby 5 pseudo markers, located in linkage groups 8, 13, and 16. Although it can be more delicately accurate results if the bivariate phenotype information were fixed by extra environment effects, those markers are valuable to be candidates for representing the resistance of VHSV in terms of flounder.
Figures & Tables
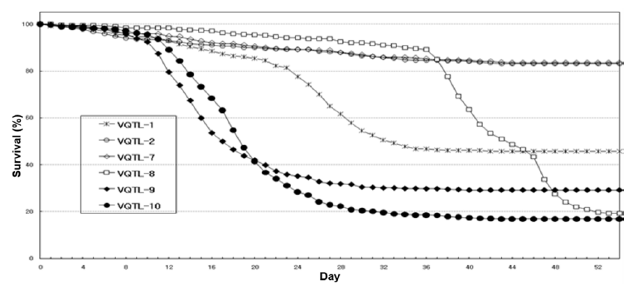
Fig. 1. Survival curves according to VHSV artificial infection experiments


