Introduction
The Thoroughbred breed originated in the early 1700s, by the crossbreeding of Arabian stallions and indigenous mares in England. Modern males originate from one of three stallions (Godolphin Arabian, Byerley Turk, and Darley Arabian) and modern females originate from one of approximately 70 foundation mares (Willett 1970). The aerobic capacity of the Thoroughbred horse is excellent unlike that of other species of similar size because Thoroughbreds have a high aerobic capacity and maximum oxygen uptake (Jones et al. 1989; Young et al. 2002). These specific characteristics have been enhanced by the intense artificial selection of sequence variations that contribute to special racing performance (Gu et al. 2009).
The key achievements of the Horse Genome Project are the complete sequencing of the Thoroughbred horse (EquCab 2.0) and the result of 1,162,753 single nucleotide polymorphisms (SNPs) across different breeds (Wade et al. 2009). The SNP of the myostatin gene (MSTN) (g.66493737C>T) has been associated with muscle hypertrophy in mammals and racing performance in horses (Hill E et al. 2010; Hill EW et al. 2010).
SNPs located in the gene coding region are classified into two types: synonymous SNPs that cause no change to amino acid metabolites and non-synonymous SNPs (nsSNPs) that affects the protein structure and molecular function. However, approximately 90% of nsSNPs cause a change in amino acid metabolites and protein function (Wang and Moult 2001). SNPs located in the coding region cause mutations in the amino acid sequence. Several studies show that the effects of amino acid allelic variants on protein structure and function can be predicted by the analysis of multiple sequence alignments and protein 3D structures (Chasman and Adams 2001; Ng and Henikoff 2001; Sunyaev et al. 2001).
We estimated the evolutionary relationships of horses using the dielectric data of six types of mammals (horse, human, mouse, dog, pig, and cow) and horses based on the whole transcriptome analyses of Thoroughbred horses before and after exercise (Park et al. 2012; Kim et al. 2013). As a result, evolutionary selected breed-specific genes, including gap junction protein alpha 4 (GJA4) and a novel SNP, were discovered. GJA4 delivers an electrical signal between cells directly through the gap junction composed of connexins (White and Paul 1999).
In this study, we focused on the SNP structure characterization of GJA4 in Thoroughbreds. The transcription factor (TF)-binding sites in the regulatory region of GJA4 were predicted. In addition, we discovered a novel nsSNP in this gene and predicted its influence on protein structure.
Material and Methods
Experimental samples
Blood samples were collected from 87 domestic Thoroughbred racehorses that had run a race at the Seoul Lets Run Park. To extract genomic DNA, 900 mL of red blood cell (RBC) lysis solution was added to 300 mL of blood, processed for 3 min, and centrifuged at 15,000 rpm for 30 s. The supernatant was removed, 300 mL of cell lysis solution and 100 mL of protein precipitation solution were added, and the solution was processed and mixed thoroughly. The DNA solution layer was collected by centrifuging the solution at 15,000 rpm for 5 min, and the supernatant was added to 300 mL of isopropanol and shaken slowly. The resulting solution was centrifuged at 15,000 rpm for 10 min and the supernatant was removed. Ethanol (500 mL) was added to the supernatant, the solution was shaken until it became clear, and centrifuged at 15,000 rpm for 3 min. DNA was extracted by volatilizing and removing ethanol (protocol number: PNU-2017-1553).
Polymerase chain reaction (PCR) analysis
NCBI (http://www.ncbi.nlm.nih.gov) and Ensembl Genome Browser (www.ensembl.org) were used to retrieve gene sequence information. The primers used to detect SNPs were synthesized using PRIMER3 software (http://bioinfo.ut.ee/primer3-0.4.0/), and the synthesized primers included GJA4 Primer F (3′-CCGGCTTCTAGCACACTCTT-5′) and R (3′-TACCTGGGCCACGTCATTTA-5′). To determine the genotype of GJA4 SNPs, PCR was used on the genomic DNA of racehorses using the following conditions: initial denaturation at 94°C for 10 min; 40 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, extension at 72°C for 30 s, and final extension at 72°C for 10 min. PCR products were separated in a 1.5% SeaKem® LE agarose gel (Lonza, Rockland, USA), detected under UV light and subjected to Sanger sequencing for confirmation after cloning. Cloning of PCR products was carried out using a pGEM®-T Easy Cloning Vector System (Promega), and each gene sequence was confirmed by Sanger sequencing. SNPs were examined by comparing the gene sequence obtained after sequencing using those obtained from a BLAST search (National Center for Biotechnology Information, Bethesda, MD, USA).
TF-binding site prediction
TF binding sites were predicted with ALGGEN PROMO v8.3 (http://alggen.lsi.upc.es).
Protein structure prediction
To investigate the amino acid sequence changes of GJA4 according to SNP, protein structure prediction analysis was performed and the following tools were used: Sorting Intolerant from Tolerant (SIFT) (http://sift.jcvi.org/), Polymorphism Phenotyping v2 (Polyphen-2) (http://genetics.bwh.harvard.edu/pph2), Protein Variation Effect Analyzer (PROVEAN) (http://provean.jcvi.org/), and I-Mutant 2.0 (http://folding.biofold.org/i-mutant/i--mutant2.0.html). Each tool was used to predict the effect of amino acid sequence substitution on the biological function of the protein and the change in the structure and stability of the protein.
3D structure analysis
Phyre2 (http://www.sbg.bio.ic.ac.uk/~phyre2) was used to predict the tertiary structure of proteins. The tridimensional structure was analyzed, and the change of structure elements was validated by referring to the structural classification method of proteins database and data in the protein data bank. Chimera v1.11 was used to confirm the sequence and comparison between protein structures according to SNP sequences.
Results and Discussion
Expression pattern of horse GJA4 and GJA4 associated genes
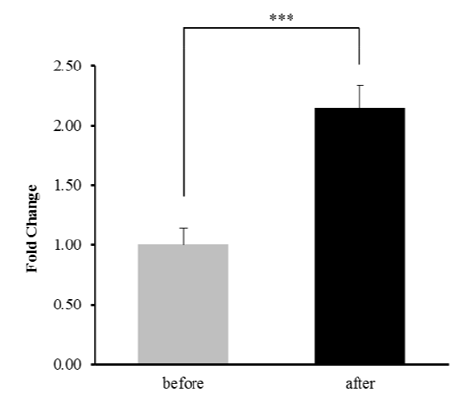
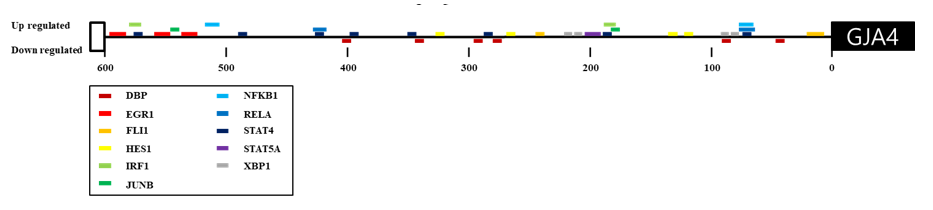
In our previous studies, we identified GJA4 as a differentially expressed gene in response to exercise and a selection signature gene related to athletic adaptation of Thoroughbred muscle (Park et al. 2012; Kim et al. 2013). From these results, the expression of GJA4 in the muscle was 2.14-fold higher after exercise than before (Figure 1). To explain the expression patterns, 164 TF binding sites were predicted in the 600 bp upstream region of the GJA4 using PROMO (data not shown). When the predicted TF genes were matched to the differentially expressed genes in response to exercise, early growth response 1 (EGR1), interferon regulatory factor 1 (IRF1), JUNB, nuclear factor kappa B (NFKB), and X-box binding protein 1 (XBP1) genes were up-regulated, and the D site of albumin promoter (DBP) gene was down-regulated after exercise (Figure 2 and Table 1).
Interferon regulatory factor 1 (IRF1) is involved in most pattern recognition receptor (PRR) signaling events, including the immune activation capacity that links innate and adaptive immunity. PRR is a receptor that recognizes the pathogenic molecule patterns (PAMPs) that cause congenital immune responses (Akira et al. 2006; Tamura et al. 2008). The transcription factor nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) plays an important role in many cellular processes and is involved in anti-inflammation (Pereira and Oakley 2008). NF-kB is activated by proinflammatory cytokines (Moynagh et al. 1993). Cytokines are intercellular signaling proteins that affect target cells and are involved in immunologically dependent inflammatory reactions. Exercise-induced skeletal muscle damage causes acute inflammatory responses and activates the muscle fiber regeneration processes. Various cytokines regulate in vivo muscle inflammation and muscle fiber regeneration (Cannon and Pierre 1998; Oppenheim 2001). Expression of zinc finger protein early growth response 1 (EGR1) induces the expression of various environmental signals including growth factors, hormones, and neurotransmitters. EGR1 is involved in the regulation of growth and differentiation (O'Donovan et al. 1999; Thiel et al. 2000), is an important regulator of pathological cardiac growth, and plays a pivotal role in the coordinated transcription of several inflammatory and coagulation factor genes, including those related to the pathogenesis of atherosclerosis and restenosis after vascular injury (Khachigian et al. 1996; Yan et al. 2000; Buitrago et al. 2005).
The GJA4 polymorphism (C1019T) is closely related to human atherosclerosis and myocardial infarction (Wong et al. 2006). The aerobic capacity of Thoroughbred horses is superior, and maximum oxygen uptake is associated with athletic performance in horses (Jones et al. 1989; Harkins et al. 1993; Gauvreau et al. 1995; Young et al. 2002). GJA4 expression is increased at the heart muscle of the Goto-Kakizaki type 2 diabetic rat after exercise (Salem et al. 2013). Therefore, we concluded that GJA4 could be an important regulator of cardiac metabolism and inflammatory responses that were caused by exercise.
Protein prediction analysis between SNPs
In GJA4, the existence of an SNP that changed C to G on the 22385534th sequence of chromosome 2, and three genotype alleles (CC, CG, and GG) were observed. Genotypes were analyzed in 87 Thoroughbred racehorses. The frequency was 82.76% for CC type, 14.94% for CG type, and 2.30% for GG type. As a result of predicting the structure of proteins created according to amino acid replacement owing to SNPs, it was demonstrated that the final product varies according to the allele (Figure 3). In GJA4, amino acid replacement from arginine to proline occurred in amino acid sequence no. 132. To predict whether the mutation of nsSNP influences the function of the protein, protein structure prediction analyses were carried out. Four tools were used to analyze the amino acid sequences substituted by the reference amino acid sequence and the SNP type. SIFT is a tool that predicts whether SNPs affect protein function by using amino acid sequence homology. PolyPhen-2 is a tool that predicts the structure and function of an SNP based on sequence information of amino acid substitutions of nsSNPs. PROVEAN predicts all protein sequence changes such as indel and amino acid sequence substitutions. Finally, I-mutant is a tool that predicts the protein stability free-energy change of nsSNPs. Only Polyphen-2 showed that SNPs affect the protein (Table 2). After 3D-tertiary structure analysis, we observed that a beta-strand changed to an alpha-helix and protein structure changed owing to the influence of SNPs (Figure 4A and B). This indicated that nsSNPs influenced the structure of proteins.
In conclusion, we proposed to correlate the induction of GJA4 and the regulation of inflammatory responses caused by muscle damages and cardiac metabolism in horse exercise. We discovered a novel nsSNP of horse GJA4 that affected protein structure. This study contributes to future research into the exercise ability of horses.