Abstract
With rapid advances in next-generation sequencing technology, previously released reference genomes are being upgraded, and new genome assemblies for novel organisms are being published. In recent updates of the bovine reference genome, changes in single-nucleotide polymorphism (SNP) position and gene location occurred. The BovineSNP50 BeadChip series and Hanwoo SNP50K BeadChip are based on the older version of the bovine reference assembly
Figures & Tables
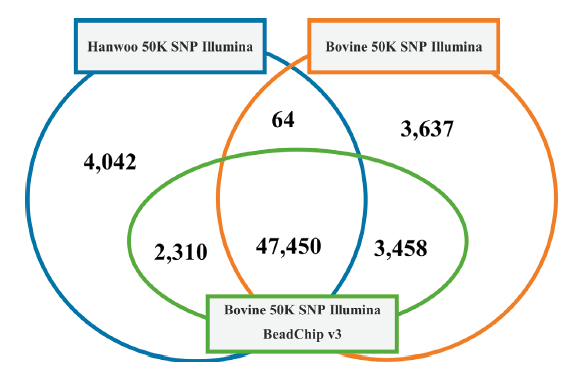
Fig. 1. Structure of Hanwoo 50K SNP Illumina BeadChip v1, Bovine 50K SNP Illumina BeadChip v2 and Bovine 50K SNP Illumina BeadChip v3. 47,450 SNPs were identified as common among the three panels (Hanwoo v1, Bovine v2, and Bovine v3). Each pair of panels shares SNPs ranged from 64 to 3,458. Hanwoo v1 and Bovine v2 do not shared 4,042 and 3,637, respectively, with the other panels.


