INTRODUCTION
Dogs (Canis lupus familiaris) hold the unique position among domestic animals as they display the greatest levels of phenotypic diversity on the planet (Clutton-Brock and Serpell 1995). More than 450 breeds are recognized from multiple sections of the globe; and are characterized by distinct morphologic and behavioral phenotypes as the result of limited gene flow and generations of intense selection over the last 300 years (Ostrander and Kruglyak 2000). Genetic studies on domestic dogs have focused heavily on established breeds with a few exceptions including a recent population genetics study on New Guinea singing dogs (Surbakti et al. 2020).
Korean Donggyeong dogs (KDG) represent one of the indigenous Korean dog breeds originating from southeast Korea (Gyeongju) over 1,000 years ago. The KDG breed is uniquely characterized by a short tail which had given rise to incorrect and negative social perceptions. They are also distinctive among native Korean dogs, e.g. Jindo (KJD) and Poongsan (KPS) dogs, and have the oldest recorded history evidenced by their numerous appearance in historical documents. Unfortunately, KDG have experienced the dramatic loss of genetic diversity (Choi 2010).
KDG, along with KJD and KPS originated in the Asian spitz clade (Choi et al. 2017, Parker et al. 2017); however, specific breed composition and hybridization in the history of Asian breeds remain uncertain. Descending from a very small founder population, KDG have experienced negative selection based on the wrong social perceptions, which have led to the population size decline to less than 500 dogs alive today, mainly bred for conservation efforts. Hence, the genomic data available today may represent a significant evolutionary unit essential for guiding future conservation and management and provide comprehensive insight into dog domestication in Korea. This study specifically attempts to assess the hybridization between KDG and 168 representative purebreds from around the world.
MATERIALS & METHODS
Genotype Data and Haplotype Sharing Analysis
The raw data files for the single nucleotide polymorphism (SNP) genotype arrays reported in this paper are retrieved from previous publications (Choi, et al. 2017). The combined data included 146,300 SNPs in 1,545 individuals from 169 breeds. The genotype data were phased to infer the haplotype sharing using the Beagle v4.1 (Browning and Browning 2007) ibd option with sliding windows of 1,000 SNPs and 50-SNP overlap parameters. The identical-by-state haplotypes between every pair of dogs were predicted to be IBD, which are based on the excessive length and strong sharing with a LOD score greater than 3.0, following the previously published method (Parker, et al. 2017). The inferred IBD haplotypes for each pair were summed across the whole genome to determine the recent breed ancestry.
Population Structure Analysis
We used the genome-wide complex trait analysis (GCTA) tool for PCA (Yang et al. 2011) which implements EIGENSTRAT (Price et al. 2006) to estimate eigenvectors, incorporating genotype data from all samples from breeds of the Asian/spitz clade. A second PCA was performed including only dogs originating from Asia: KJD, KPS, Akita, Shiba Inu, Chow Chow, Tibetan Mastiff, and Chinese Shar-pei. The inbreeding coefficient (F) and nucleotide diversity (in windows of 10 Mb) statistics for each breed were estimated using VCFtools (v0.1.13) (Danecek et al. 2011).
RESULTS & DISCUSSION
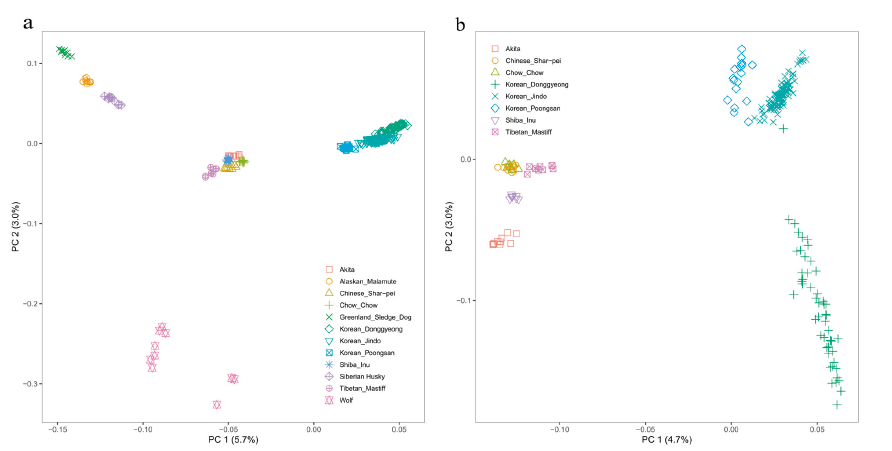
Leveraging the publicly available data, we created a dataset of 1,545 dogs from 169 breeds including Korean indigenous breeds, KDG, KJD, and KPS (Supplementary Table S1). We first undertook Principal Component Analysis (PCA) of the autosomal genotype data from KDG and Asian and spitz breeds (Figure 1), which presented the global patterns of genetic structure. The analysis revealed clear structures and separations as samples from the same breed including KDG clustered together. The samples showed no evidence of admixture with each other. The wolf samples clearly constitute a distinct group, and the split within the population is due to independent Korean lineage formation stemming from a bottleneck. To further examine genetic relationships among Asian breeds, we conducted an independent PCA on KDG and Asian breeds, where PC 1 provided evidence that Korean indigenous breeds and Chinese and Japanese breeds are genetically discriminated. The wide distribution of Korean indigenous dogs, specifically DG, indicates the higher level of heterogeneity in this population than in other established Asian breeds.
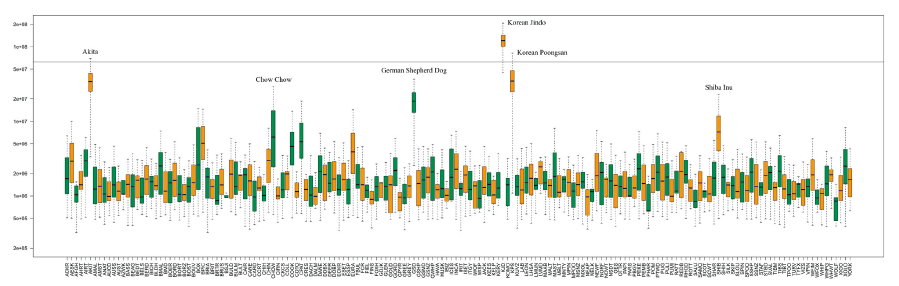
To capture a holistic view of the genetic and breed composition of KDG in comparison with all publicly available purebreds, we assessed possible hybridization using identical-by-descent (IBD) haplotype sharing. As expected, other indigenous Korean dogs such as KJD and KPS showed the significant amounts of shared haplotypes, reflecting the geographical and historical relatedness of these breeds. The Japanese breeds Akita and Shiba Inu and a Chinese breed Chow Chow also showed strong haplotype sharing with the KDG. The results corroborate with the ancient literatures, which state that most indigenous Korean dogs migrated to Japan and may have contributed to the genetic diversity of ancestral Japanese dog populations (Choi 2010). Finally, a European herding breed, German Shepherd dogs also indicated the unexpected substantial hybridization with KDG.
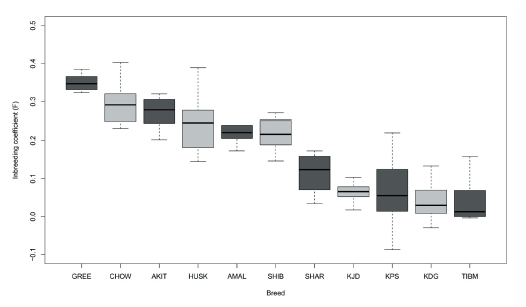
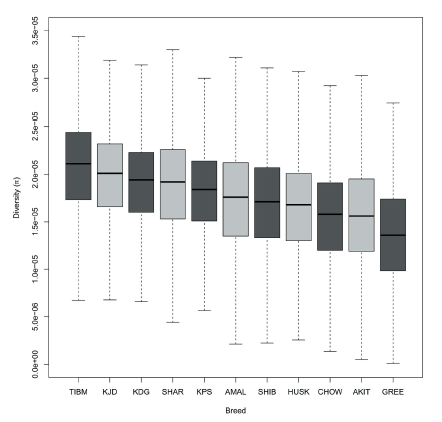
We next examined the inbreeding coefficient (F) in the KDG and Asian/spitz breeds (Figure 3). Of the eleven breeds estimated, Akita showed the one of the highest inbreeding coefficients, illuminating the strong and concerted closed breeding practices, as Akita was recognized as an official breed by the American Kennel Club (AKC) in 1972. On the other hand, Tibetan Mastiff had the lowest inbreeding coefficients, suggesting a relatively greater genetic diversity within the population compared to other established breeds. KDG were ranked tenth from eleven breeds tested and lower than both KJD and KPS. This may indicate that the current low current population size of KDG could be misleading in terms of the genetic diversity due to the limited efforts of controlled and selective breeding. This result was also supported by the nucleotide diversity analysis by each breed, which showed that KDG had a relatively high genome-wide diversity level compared to purebreds (Figure 4). Nucleotide diversity (π) is defined as the average occurrence of nucleotide discrepancies per site between any two randomly chosen DNA sequences to estimate the degree of polymorphism within a population (Nei and Li 1979). The early ancestral population of KDG were not subject to intensive and closed breeding schemes and may have contributed to the rich nature of the standing genetic variation in the KDG population today.
The genomes of domestic dog breeds offer a lens through which to view genetic variation and understand evolutionary forces on an evolving population. Given the evidence of both drastic population decline and restoration, KDG represent a valuable resource for population genetic studies. Our results revealed KDG shared significant proportion of haplotypes across genome with geographically close populations including indigenous Korean breeds and a Japanese breed Akita. Finally, we observed a relatively high level of standing genetic variation in KDG when compared to closely related purebreds, although the negative selective pressure over time may have costed a substantial loss of genetic diversity. This study shows the value of comparative genomics in disentangling signals of the admixture and demographic footprints in the population history.