Introduction
Hanwoo (Korean cattle) has a long history used as a draft animal before being the most profitable cattle as meat-type one since 1960s along with the development of the Korean economy (S.-H. Lee et al., 2014). The cherished role in the food industry of Hanwoo is due to its palatability, which is performed by several characteristics such as tenderness, juiciness, or flavor and determined by the consumers with the complexity of processes that occur while chewing (Caine et al., 2003). As one of the most important features for the quality of meat, research has been conducted for increasing the satisfaction of the customers about tenderness. However, tenderness is highly variable as it is affected subjectively and objectively by several factors.
A considerable number of methods have been developed to predict meat tenderness, which is mainly sensorial or instrumental. While the sensory evaluations are supposed to be expensive, take a lot of time and labor, instrumental methods are established to measure the meat tenderness (Caine et al., 2003). Warner – Bratzler Shear Force (WBSF) values are the most widely used, which is the force required to cut through a piece of meat (Wheeler et al., 1997). The range indicated by the customers from a study of “tough” meat is larger than 52.68 N and the counterpart is lower than 42.87 N (Destefanis et al., 2008), explaining a substantial proportion of variation in sensory assessment of beef tenderness.
Moreover, tenderness is a quantitative trait, controlled by many genes and affected by environmental effects. Besides the amount of marbling, the extent of the proteolysis of cytoskeletal, cytoskeletal-associated proteins, and extracellular matrix-associated proteins postmortem, the amount and solubility of the connective tissue is one of the most important factors determining the meat tenderness. 12 genes was published previously that related to the amount and solubility of the connective tissue, including CCR4-NOT Transcription Complex Subunit 3 (CNOT3), Osteoclast Associated, Immunoglobulin-Like Receptor (OSCAR), NLR Family Pyrin Domain Containing 5 (NLRP5), which are associated with the connective tissue score (Leal-Gutiérrez & Mateescu, 2019). Moreover, the genes PIBF1, which encodes a cytoskeletal pericentriolar protein and ITBG3 encoding a transmembrane protein also have a connection to the palatability-related traits, while collagen genes COL11A1, COL1A , COL24A1, COL28A, COL2A1, COL4A3, and COL6A3 are also in the association with tenderness (Leal-Gutiérrez & Mateescu, 2019).
Additionally, the candidate gene approach is one of the initiations of association studies. This kind of study has been focusing on genes whose functions are already known or related to traits, through previous studies. It is considered as the validation of the orientated guess conducted from the genetic basis research. Starting from selected target genes, whose roles were preconceptions, the polymorphisms, which are usually Single nucleotide polymorphisms (SNPs), are accessed to find out their effects on these traits. SNPs with significant effects would be used as potential biomarkers which may have prospective application in the future (Kwon & Goate, 2000).
Therefore, this association study was performed to identify the distribution of potentially functional SNPs or candidate regions associated with tenderness from 12 candidate genes collected from the previous study. SNPs results may be used as markers for this trait selection.
Materials and methods
Results
1. Animals and meat tenderness measurement
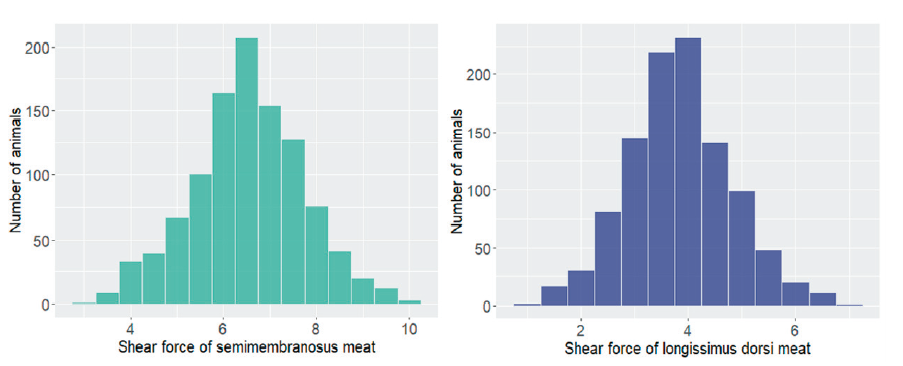
A total of 1,161 Hanwoo steers born between 2010 and 2014 at Hanwoo Experiment Station, National Institute of Animal Science (NIAS), Rural Development Administration (RDA), South Korea, were used in this study. Animal health and welfare guidelines of the Animal Care and Use Committee (NIAS) were followed (ethics committee approval number: 2015-150). Phenotypic data on WBSF of longissimus dorsi area (D_SF) and semimembranosus area (S_SF) from 1,161 animals were measured according to the method by (Wheeler et al., 2000). 2.5 cm2 thick beef steak kept in the polyethylene bags 48 hours after slaughtering was heated in water 30 minutes at 80oC until reaching 70oC itself, then stored at room temperature 30 minutes before measuring. The WBSF value, which is the maximum force required to shear each of six cores divided parallelly to the muscle fiber from each sample, will be obtained using an Instron Universal WBSF testing machine (Instron Corporation, Canton, MA) (H. J. Lee et al., 2020). The descriptive statistics of phenotypic data are shown in Table 1 and Figure 1.
|
Table 1. Descriptive statistics for Warner-Bratzler shear force (kg/0.5 inch2) of semimembranosus meat and longissimus dorsi meat 
|
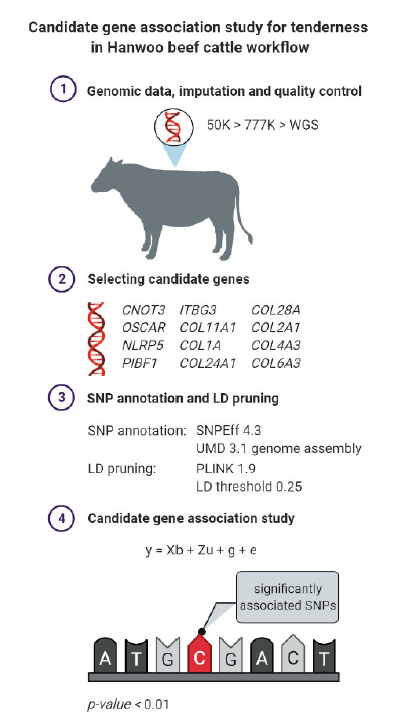
2. Genomic Data, Imputation and Quality Control
1,161 cows with phenotypic data was genotyped using the Illumina Bovine SNP50 BeadChip (50K) (Illumina, San Diego, CA, version 2) and was subsequently imputed from 50K to 777K to whole sequence using Minimac3 (Bhuiyan et al., 2018). The reference data set includes 1,295 animals genotyped with 777K SNP chip (Illumina Bovine HD Beadchip, Illumina, San Diego, CA) and 203 animals at the level whole-genome sequence. The quality control was performed with the control thresholds: minor allele frequency (MAF) >0.01, missingness rate per marker < 0.01, and Hardy-Weinberg equilibrium (HWE) p-value <0.0001 after each step (H. J. Lee et al., 2020). 38,623 SNPs survived among 54,931 SNPs after QC for the genotyped data of 1161 cows at 50K level and 618,818 SNPs was given as the result of QC for imputation from 50K to 777K. Finally, 11,227,701 SNPs were the result of imputation to whole sequence level and were used for analysis in this study.
3. SNP Annotation and LD pruning
Candidate genes were selected from a previous study about meat palatability, in which connective tissue contributing to meat tenderness was shown to be in association with 12 genes (Leal-Gutiérrez & Mateescu, 2019). These genes (CNOT3, OSCAR, NLRP5, PIBF1, ITBG3, COL11A1, COL1A, COL24A1, COL28A, COL2A1, COL4A3, and COL6A3) were used for analysis in this study. UMD 3.1 genome assembly was used as reference sequence for determining the physical position of the imputed SNPs (Bhuiyan et al., 2018). SNPEff software 4.3 was used to perform the annotation (Bhuiyan et al., 2018). The annotated SNPs were partitioned into groups based on their genomic location. SNPs obtained was pruned using PLINK 1.9 (Purcell et al., 2007) based on linkage disequilibrium (LD), with the LD threshold 0.25, which means if the pairwise LD is greater than 0.25, one SNP of the pair would be pruned, remaining the SNP with the largest minor allele frequency (MAF).
4. Candidate gene association study
Association study was performed using a linear mixed model. a linear mixed model with GCTA 1.93.2 (Yang et al., 2011). The statistical model for the phenotypes (D_SF and S_SF) can be written as:
y = Xb + Zu + g + e
Where y is an n x 1 vector of phenotypic values with n being the sample size (n = 1161) and variance var(y) = V = Gσg2 + Iσe2 where G is the genetic relationship matrix (GRM) estimated using whole-genome SNPs with GREML module of GCTA and I is n x n identity matrix; b is the vector of fixed effects, including growing location (district, ward), time of slaughtering (year, month), time of birth (year, month), and age (days), u is the vector of SNP effects with u ~ N(0, Iσu2); g is the n x 1 vector of the total genetic effects with g ~ N(0, Aσg2); e is the vector of random residual effects, which is the part of variance not accounted for by the variables in this model with e ~ N(0, Iσe2).
Finally, significant SNPs associated with the phenotype were obtained by using 0.01 as the significant level. SNP fitted to the model having the p-value <0.01 will be in a significant association with the trait. The analysis workflow is shown in Figure 2.
1. Annotation and LD pruning
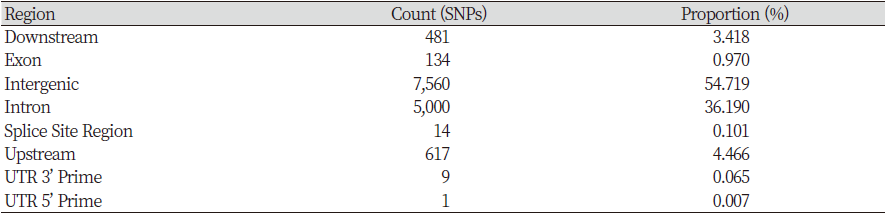
We conducted SNPs annotation using SNPEff 4.3 (Cingolani et al., 2012) to find the genetic variants for the 12 candidate genes and obtained 12,319 SNPs from these genes, belonging to 9 types of variants or 8 types of regions (Table 2). The largest number of SNPs was 7,560, in the intergenic region, consists of 54.72% of the total. The second significant number was 5,000 SNPs, makeup 36.19% of the data and was in the intron region. The upstream and downstream regions also have 617 and 481 SNPs detected, accounting for 4.47% and 3.48%, respectively.
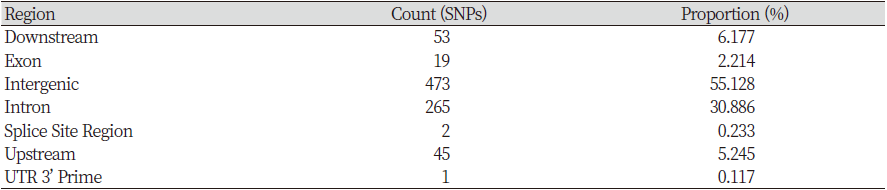
For LD pruning, a threshold of 0.25 was applied, resulting in 730 SNPs, belonging to 8 categories of variant or 7 regions (Table 3). The highest number of SNPs was still in the intergenic and intron regions, comprising 55.13% and 30.89% of the total, which are severally 473 and 265 SNPs.
2. Candidate gene association study
Among a total of 730 SNPs, significant 10 and 7 SNPs for S_SF and D_SF respectively were detected from association study and these SNPs were passed the significant threshold (p-value < 0.01) (Table 4, Table 5, and Figure 3). According to the analysis result for S_SF, 10 SNPs are found to be in significant effects in total, including 2, intergenic and intron, SNPs on chromosome 2 related to the COL4A3 and COL4A4 genes; 1 and 3 intergenic SNPs related to the genes COL11A1 and COL6A3 on the chromosome 3, respectively. 4 SNPs which are intron variances were found to be in association with the PIBF1 gene on the chromosome 12. The analysis has also shown the result that 7 SNPs are significantly associated with D_SF, with 3 SNPs located on chromosome 3 in the intergenic region related to the gene COL6A3, 2 SNPs to be linked with the gene PIFB1 on chromosome 12 and the witness of 2 intergenic SNPs on chromosome 19 in connecting with the gene COL1A1.
|
Table 4. Significant SNPs found from 12 candidate genes associated with WBSF of semimembranosus meat under the threshold of significance p -value < 0.01 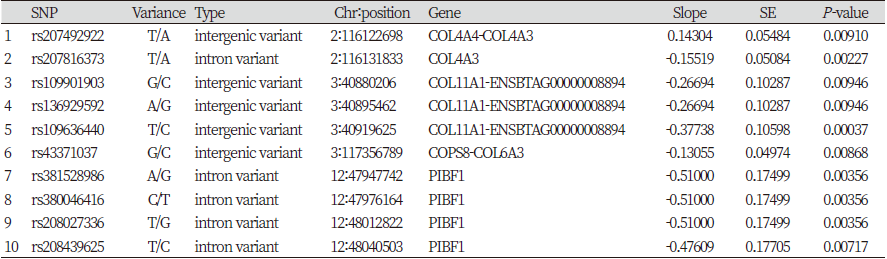
|
|
Table 5. Significant SNPs found from 12 candidate genes associated with WBSF of longissimus dorsi meat under the threshold of significance p -value < 0.01 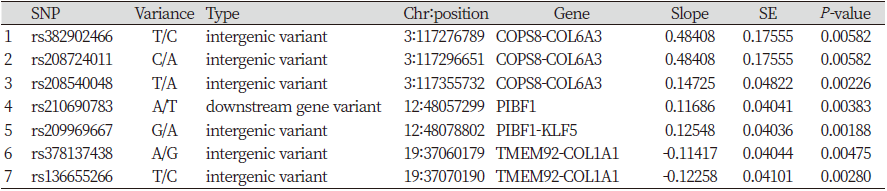
|
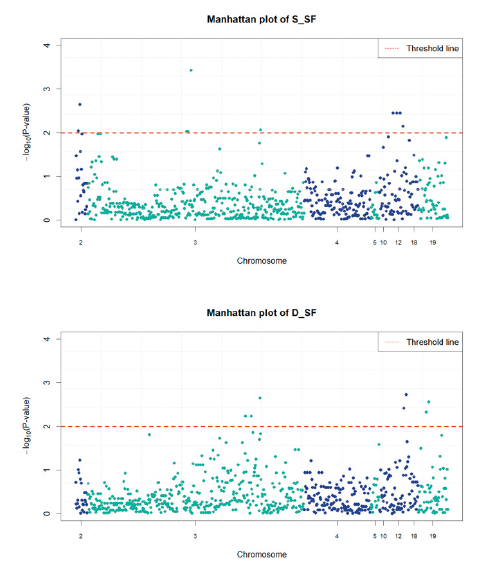
Fig. 3. Manhattan plot of the association study for Warner-Bratzler shear force of semimembranosus meat (above) and longissimus dorsi meat (below). The y-axis shows the −log10(p-value) of each SNP and the x-axis is the SNP index. The red line represents the threshold of significance (p-value = 0.01).
Discussion
The palatability, which is the factor making the Hanwoo be the most profitable cattle in Korea, is built from several features. Tenderness is one of those. As reported in previous study, the contractile proteins and connective tissue are the two components determining the meat tenderness (MARSH, 1977). Being highly important contribution, connective tissue related factors, composed of collagen, elastin and reticulin (Weston et al., 2002), have been emphasized in many studies for meat tenderness. While elastin and reticulin do not play essential roles in the toughness of meat, collagen does significantly (Weston et al., 2002). Therefore, detecting SNPs in association with collagen genes, which are major candidate genes in this study, is reasonable for discovery of meat tenderness biomarkers.
We extracted SNPs from 12 genes from a previous study to be the candidates of this research (Leal-Gutiérrez & Mateescu, 2019). As mentioned above, we found out 10 significant SNPs for S_SF and 7 SNPs for D_SF. Most of them are the SNPs being related to the gene belonging to the collagen family that we found to be interesting result in this study. Collagen have been reported to contribute in meat tenderness and textures in beef cattle through functioning in connective tissues, and the alternation of its performance plays an important role in delivering desired tenderness (Weston et al., 2002). Collagen is one of three proteins composing the connective tissue, beside elastin and reticulin, appearing ubiquitously in the muscle and being the most abounding proteins in mammals (Ricard-Blum, 2011). There are 28 members in the collagen family, each has the tropocollagen as the basic unit, made up of 3 α-chains in their structure (Shoulders & Raines, 2009). Our result has shown that a remarkable number of significant SNPs associated to tenderness is related to collagen type I, IV, VI and XI genes.
Collagen I is a fibrillar collagen, which is the most abundant intramuscular collagen of meat (Weston et al., 2002). Composing of 2 α1 chains and 1 α2 chain, type I collagen molecule is heterotrimer most of the time and hardly to be the homotrimer of 3 α1. With the length of about 300 nm, each chain of the molecule comprises more than 1,000 nucleotides, including 3 domains: the N-telopeptide, the central triple helical domain and the C-telopeptide (Henriksen & Karsdal, 2016). Due to its large amount and ubiquity, the biomarkers of this type of collagen, which includes synthesis and degradation biomarkers, have been well-documented. While the synthesis biomarkers are usually used in clinical purposes, the degradation markers are related to the crosslinks (Henriksen & Karsdal, 2016), which are available in the fibrous collagen and are significantly in association with the meat tenderness (Weston et al., 2002).
Collagen XI is also a fibrillar collagen, which, along with type I, acts as the toughness causing factor of the meat (Weston et al., 2002). Collagen XI, as a member of collagen family, also be omnipresent in the body (Luo & Karsdal, 2016). Therefore, the appearance of it causes the meat to be tough, which is unfavorable. It was reported that the collagen XI cannot be broken physically through cooking process (Weston et al., 2002). Thus, it suggests to the way to meddle it in meat may be through regulation of its genes or through selection. SNPs in the intergenic regions that we found may have a potential chance to be markers for the regulation, or intron variants may also be the marker for selection for the collagen XI genes specifically or tenderness traits generally, causing the advantageous diversification for the quality of meat as an expectation. Although it plays a very important role in the meat tenderness, it has been reported to be no biomarker available for Collagen XI until 2016 (Luo & Karsdal, 2016).
One of the minor types of filamentous collagen, collagen VI, is also found in muscle. Similar to the other members of the family, collagen VI is triple helix, with the anti-parallel alignment forming between chains. There is still no clear information about the role of this collagen type in the meat texture (Weston et al., 2002).
Although the collagen IV is not a fibrous collagen, it is still available in the muscle, being the only non-fibrous one there (Weston et al., 2002). With 6 types of the primary structure, it forms 3 different triple helical organizations, including: α1α1α2, α3α4α5 and α5α5α6 before forming the lattice-shaped network (Sand et al., 2019). The type IV collagen is the backbone of the cell membrane, functioning in forming network and compartmentalizing the tissues. It is considerably related to most of the connective tissue diseases (Sand et al., 2019) and clearly plays an important role in the texture differences of meat (Weston et al., 2002). Hence, several biomarkers for this type of collagen have been well-documented (Sand et al., 2019).
A threshold of 0.25 was set for LD pruning. Obviously, LD pruning bring an advantage in pre-analysis as increasing the signal sensitivity and reduce the type I error (false positive rate) of the result as compared to no pruning at all. On the other hand, it was reported that an extremely low threshold may cause a drawback to the statistical power of the association detection (Joiret et al., 2019). However, in this study, since the functions of those genes have been introduced clearly, SNPs associated with them may be highly biased. The threshold of 0.25 was used as an appropriate parameter to detect more exact causal variants.
Conclusion
10 SNPs for semimembranosus area (S_SF) tenderness and 7 SNPs for longissimus dorsi area (D_SF) one in Hanwoo beef cattle, almost of which are in connection with collagen family genes, were detected through candidate gene association study using linear mixed model. This study hopefully provides effective genetic markers for meat tenderness trait in Hanwoo cattle.
Acknowledgement
This research funded by Research Funding (2020-0534-01) of Chungnam National University and this work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (321082-3)