Abstract
The genetic diversity of the major histocompatibility complex (MHC) molecules of pigs has not been thoroughly characterized. In this study, we successfully typed SLA-2, a pig MHC class I gene, from 10 pigs of five different breeds using a high-resolution typing method. We identified a total of 14 SLA-2 alleles including 2 previously unreported alleles and reported to IPD-MHC SLA database. These new alleles showed 1 and 29 nucleotide differences from the closely related alleles, SLA-2*10:01 and SLA-2*13:02, respectively. The information of SLA new alleles should enrich our understanding the diversity of MHC diversity in pigs.
Figures & Tables
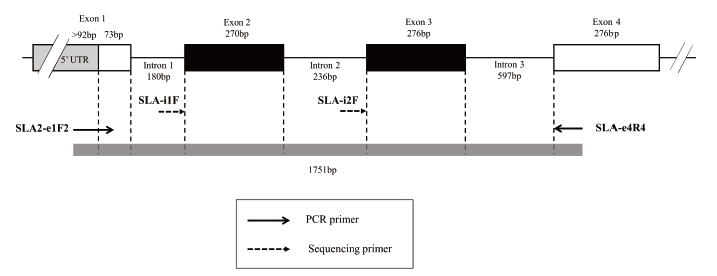
Fig. 1. General overview of the genomic sequence-based SLA-2 genotyping method. The diagram shows the location of each primer used for PCR amplification and sequencing. The sizes (bp) of introns, exons, and PCR products are indicated.


