Abstract
Genomic information is now useful to identify quantitative trait loci (QTL) which is associated with economic traits as well as to predict genetic potential for individual in Animal Breeding Industry. Especially, genomic BLUP (gBLUP) is one of the useful model to estimate genomic breeding value (gEBV) with genomic relationship matrix (GRM). Genomic relationship matrix will be estimated from genomic information such as single nucleotide polymorphism (SNP) which is similar to numeric relationship matrix estimated from pedigree information used in traditional BLUP. The matrix defines the genetic covariance between individuals based on observed similarity using genomic information, rather than on expected genetic similarity from pedigree. Therefore, gBLUP would be given to better prediction accuracy in livestock breeding.
Acknowledgements
본 연구논문은 농촌진흥청 차세대 바이오그린21사업(PJ011842012017)으로부터 연구비를 지원받아 수행하였습니다. 연구비 지원에 감사드립니다.
Figures & Tables
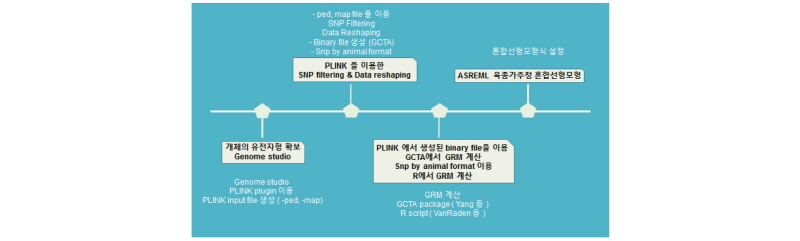
Flowchart for estimating of genomic estimated breeding value (gEBV) in ASREML


