Introduction
The history of cattle production in the Korean Peninsula dates back to over 5000 years (Lee et al., 2014; Payne and Hodges, 1997). Analysis using mtDNA showed that Korean cattle carry taurine haplotype (Mannen et al., 2004). Concurrent with the mtDNA results, in our recent study, we traced little genetic influence of African and Asian indicine in Korean Hanwoo cattle (Edea et al., 2015). On the other hand, SNPs based analysis did detect the genetic influence of African taurine and indicine in Hanwoo (Decker et al., 2014), which was believed to be introgressed by European cattle via the Silk Road (Christian, 2000). In their microsatellite-based study of north-east Asian cattle, Kim et al. (2002) have shown that Korean cattle are much closer to Chinese cattle but distinct from Japanese Black cattle.
Based on coat color phenotypes and geographical distribution, Korean native cattle are categorized into four breeds/strains: Brown Hanwoo, Jeju Black, Heugu, and Brindle Hanwoo/Chikso (Lim et al., 2016; Suh et al., 2014). Of these the four breeds, the Brown Hanwoo is the predominant breed in terms of population size (3, 000,000) and geographical distribution, whereas the population sizes of Chikso and Heugu were estimated to be only between 300 to 400 heads (Suh et al., 2014). Therefore, the later breeds may have faced severe population bottleneck. Assessment of the possible impacts of demographic events on the landscape of genetic diversity and population structure are of particular importance for the ongoing conservation program.
During the earlier periods, Korean cattle were majorly kept for the draft, transportation, and socio-cultural purposes (Lee et al., 2014b). Driven by consumers demand for quality beef products, however, since the 1960s, cattle breeding objective has geared towards meat production (Lee et al., 2014a; Park et al., 2013a) and has resulted in a substantial genetic gain. Accordingly, the Brown Hanwoo cattle breed has been subjected to intensive selection for beef and meat quality traits (Lee et al., 2014b; Park et al., 2013). Following decades of selection, a substantial genetic gain has been achieved for carcass weight and eye muscle area (Park et al., 2013b). For instance, an annual weight gain of 8 kg in carcass weight was reported (NIAS, 2009). Similarly, yearling weight was increased from 315.54 kg to 355.06 kg during the past 13 years (Park et al, 2013). On the other hand, the two native Korean cattle breeds have not been selected for beef traits. Due to selection against certain phenotypes (e.g. coat color), the Brindle (Chikso) and Black (Heugu) Korean cattle have received less attention, as a result, their population sizes have been dramatically declined. The intensive selection of Hanwoo cattle for beef traits may have left genomic signatures of selection within genes associated with beef traits. Genomic comparison of selected (Hanwoo) and unselected (Chikso and Heugu) cattle breeds can provide some insight into the possible impacts of human-guided selection in shaping the genetic composition of the former breed group. Furthermore, it can facilitate the identification of causal variants associated with a trait of interest. In this study, we present the genome-wide genetic diversity and signatures of selection analyses in three Korean native cattle breeds exposed to different demographic events/ selective pressures.
Materials and Methods
Cattle breeds, genotyping and quality control
In this study, we sampled three native Korean cattle populations, namely Brown Hanwoo (n = 248), Chikso (Brindle Hanwoo, n = 18) and Heugu (Black Hanwoo, n = 18). To account for the effect of sample size on estimates of genetic diversity and population structure analysis, we randomly selected 18 Hanwoo samples from 248 individuals. All individuals were genotyped with the 600K Affymetrix chip, which contains a total of 630,973 autosomal SNPs. The SNP markers were screened for a call rate < 95%, minor allele frequency (MAF) < 0.05. Following applying the above quality control criteria, 354,337 autosomal SNPs were retained and used for downstream analysis.
Data analyses
Genetic diversity was estimated for each breed using PLINK (Purcell et al. 2007). Inbreeding coefficient and animal relatedness were estimated as the proportion identity-by-decent (IBD) between sample pairs within the breed as a PI_HAT value using the same package. A principal component analysis (PCA) was also performed on 354,337 SNPs using SNP and Variation Suite v8.5.0 (Golden Helix, Inc., Bozeman, MT, www.goldenhelix.com). Linkage disequilibrium (LD) was estimated between adjacent SNPs. Inter-SNP distances were binned into different categories and estimates of r 2 were averaged for each inter-SNP distance category. Effective population size (Ne) in each breed was estimated from linkage disequilibrium using SNeP tool (Barbato et al. 2015) with the default settings.
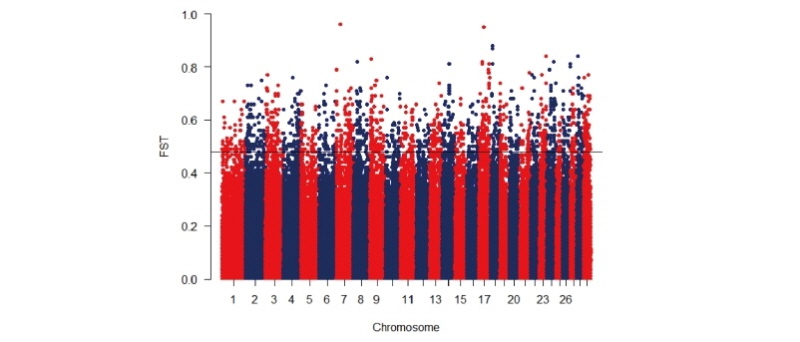
Genetic differentiation (FST) was estimated with unbiased estimator according to (Weir & Cockerham 1984) using SNP and Variation Suite v8.5.0 (Golden Helix, Inc., Bozeman, MT, www.goldenhelix.com). Based on the results of PCA and neighbor-net tree, two pairwise comparisons (Hanwoo-Chikso, and Hanwoo- Heugu) were performed to identify genomic regions that may be under selection. The highly-differentiated regions representing the top 0.5% were used to define genomic regions under divergence selection. We further performed functional annotation analysis using the web-based DAVID Bioinformatics resources (v. 6.8, https://david.ncifcrf.gov/).
Results and discussion
Genetic diversity, population structure, and demographic history
Compared to the Brown Hanwoo cattle, the two Korean cattle breeds (Chikso and Heugu) display low levels of observed heterozygosity (Table 1). These values are higher than those (0.26-0.29) estimates by (Kim et al., 2018), but far lower than the microsatellite-based values (0.67) (Suh et al., 2014). The low level of genetic diversity for Chikso and Heugu is possibily attributed to genetic drift associated with low effective population size. The levels of inbreeding coefficients were found to be -0.094, -0.127 and -0.064 for Chikso, Heugu, and Hanwoo, respectively. The estimated average relatedness as measured by the proportion of IBD was the lowest for Hanwoo (0.055). The analysis of molecular variance revealed that 10.06% (P < 0.000) of the total variability can be attributed to among populations, and almost 90% of the variation was due to within populations.
Breed-specific SNPs can be used for product traceability and breed discrimination/assignment (Negrini et al., 2009). A SNP was breed-specific when it was segregating/ polymorphic (MAF > 0.05) in the one breed but fixed in the other breeds (Czech et al., 2018). The highest number of breed-specific SNPs was found in Hanwoo and the lowest for the Chikso breed. Annotation of breed-specific SNPs showed that the SNPs were associated with 125 genes in Chikso and 116 in Heugu.
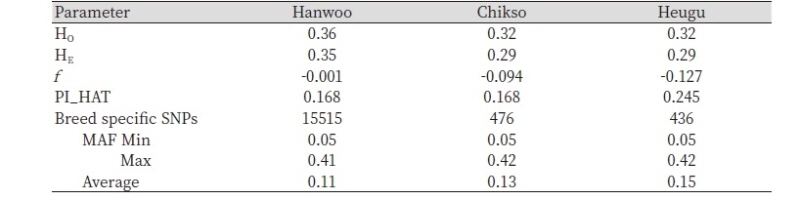
Principal components analyses nearly clustered the three breeds according to their phenotypic /coat color based discriminations. As well portrayed in Figure 1, the PC1 accounted for about 48.48% of the variation and separated Hanwoo and Chikso from the Korean Black cattle. The PC2 separated Brown Hanwoo from Brindle Hanwoo(Chikso). Consistent with PCA results, our Neighbor-joining (NJ) confirms the PCA results (Figure 2). The Heugu breed was distantly separated from the two mainland cattle breeds, possibly due to genetic drift associated with small effective population size.
LD and effective population size
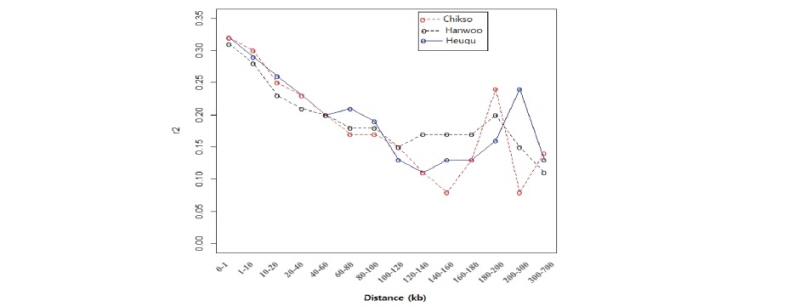
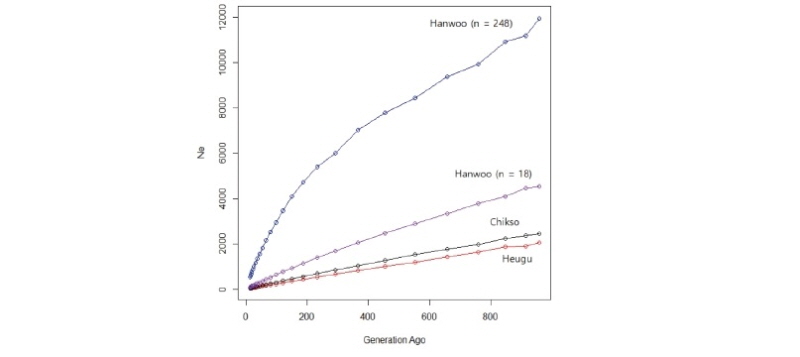
The overall mean r2 values were 0.29 ± 0.35, 0.29 ± 0.36 and 0.27 ± 0.30 in Chikso, Heugu, and Hanwoo, respectively. The value obtained in Hanwoo was higher the reported 0.17 and 0.23 using the 50K and 80K BeadChips, respectively (Edea et al., 2015). The average LD values observed in this study here were higher than those reported for the Nellore cattle breed (r2 = 0.17) (Espigolan et al., 2013). For inter-SNP marker distances of 20–40 kb, average estimated LD (r2) values for Chikso, Heugu, and Hanwoo were 0.23, 0.23 and 0.21, respectively (Figure 3). At shorter SNP distances (0-40 kb), Hanwoo had shown lower LD than the Chikso and Heugu cattle breeds. In the Hanwoo cattle, r2 values dropped from 0.28 to 0.11 when the inter-marker distance increased from 1–10 to 300–700 kb (Figure 3). The higher LD at a shorter distance in the two Korean cattle breeds could be attributed to a reduction in their effective population sizes across generations. As expected, the two Korean cattle (Chikso and Heugu) displayed smaller Ne across generations (Figure 4). These smaller Ne values obtained for Chikso and Heugu breeds are in concurrent with the observed lower within-breed genetic diversity and higher LD at shorter distances. The Ne values obtained in this study for the last 13 generations are lower than those reported for three Korean cattle populations (Kim et al., 2018). Estimates of Ne found in Hanwoo is higher than previously found for Jersey (Ne = 82), but lower than the value (Ne = 153) reported in Charolais (Edea et al., 2018).
Candidate regions under selection
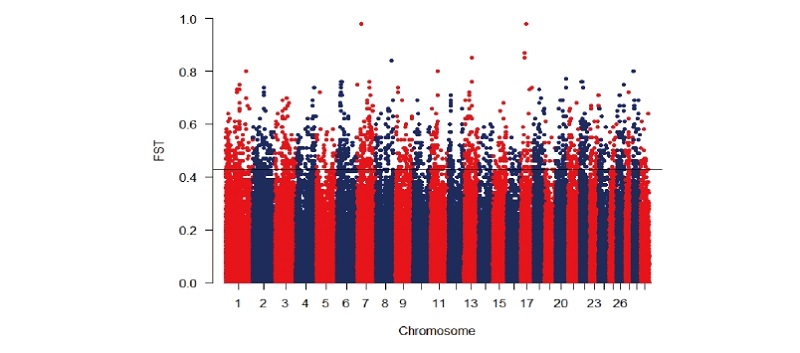
During their evolutionary histories, Korean cattle have exposed to different demographic events. The brown Hanwoo cattle have been under selection for beef traits since the 1960s, whereas the two Korean cattle breeds (Chikso and Heugu) have received less attention and are not the focus of systematic selection. The difference in demographic events and selective pressures are expected to shape the genomes of these population differently. In an attempt to capture the possible impacts of human-guided selection and/or genetic drift on the genomes of the Korean cattle breeds, we conducted a genome-wide comparative analysis on 284 animals representing three Korean cattle breeds using 354,337 SNPs. The mean genomic FST values ranged from 0.06 between Hanwoo an Heugu and to 0.13 between Chikso and Heugu, respectively. Overall, the top 0.5% FST values contained 409 and 419 genes in the Hanwoo/Chikso and Hanwoo/Heugu comparisons, respectively (Tables S1a and b). For the former breed pair comparison, the largest number of genes per chromosome were identified on BTA1, and BTA6 (31 genes). In Hanwoo-Chikso pair, the most strongly selected regions were detected on BTA6 (26.16 Mb), BTA7 (28.22 Mb), BTA8(91.27 M), and BTA17 (25.05, 35.67 Mb) containing 4 genes (MAPKSP1, LOC100126815, LOC100336402, CTNND2, respectively). For Heugu-Hanwoo pair, candidate genes associated with high FST value were noted on BTA7, 17, and 18. We further assessed selective signals between the two pairwise comparisons. Shared signals of selection were detected on BTA1 (62.17 Mb), BTA2 (34.25 Mb), BTA3(66.80-68.72 Mb), BTA5(51.27 Mb), BTA6(43.80; 60.20-60.30 Mb), BTA14 (48.57, 72.49 Mb), BTA17(25.05 Mb), BTA21 (59.41 Mb), BTA22 (37.65 Mb), BTA23(54.63 Mb), and BTA25 (38.20 Mb) containing LOC538060, IFIH1, GIPC2, LOC782689, MON2, GBA3, KLB, C6H4orf34, EXT1, CDH17, LOC100336402, PPP4R4, ATXN7, LOC540231, and LOC615412 candidate genes.
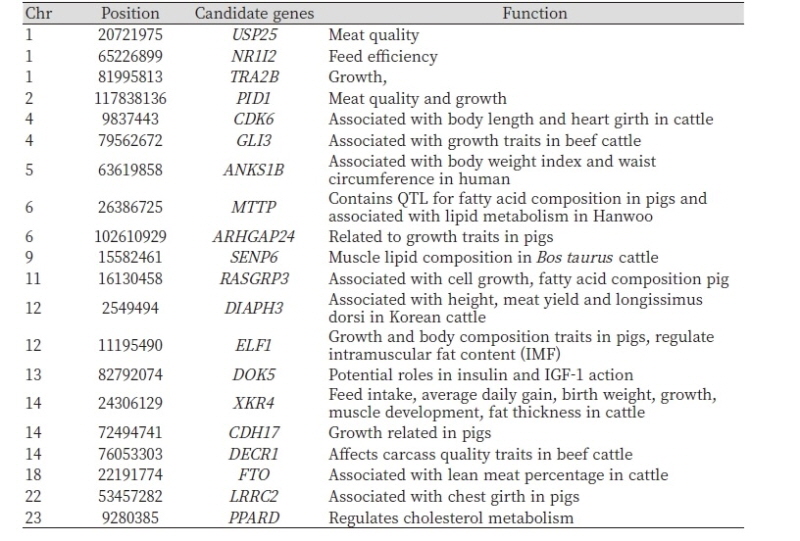
The brown Hanwoo cattle breed has been phenotypically subjected to intensive selection for meat production and quality traits including carcass weight, eye muscle area, backfat thickness, and marbling score (Park et al., 2013). Interestingly, we identified genes under selection and known to be associated with growth, stature, and carcass quality (Table 2). The XKR4 gene has been reported to be associated with backfat thickness (Porto Neto et al., 2012) feed intake and growth traits (Medeiros et al., 2017; Lindholm-Perry et al., 2102). MTTP contains QTL for fatty acid composition (Corominas et al., 2013) and associated with lipid metabolism in Hanwoo (Baik et al., 2015). The remarkable effect of the FTO gene on live weight and carcass characteristics has been extensively investigated in many studies. In their association studies using Simmental and Brown cattle, Skok et al. (2015) demonstrated that variants of FTO gene were associated with lean meat percentage, fat percentage, and live weight at slaughter. Additionally, this gene has been investigated for its effect on body mass index in humans and growth and carcass traits in pigs and cattle (Rempel et al., 2012; Fontanesi et al., 2009; Frayling et al., 2007).
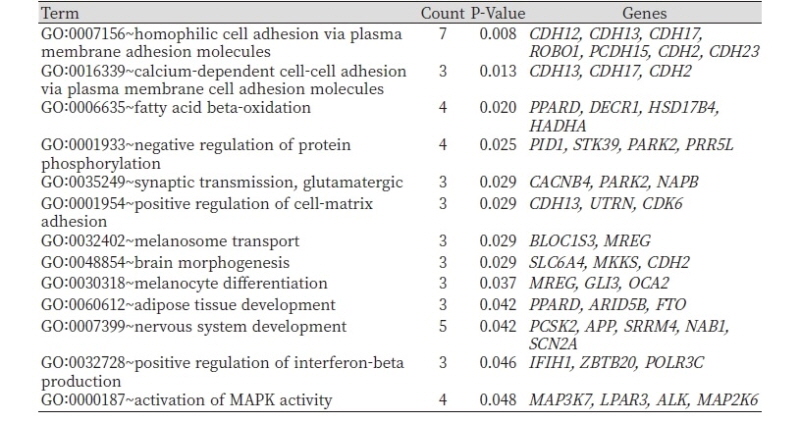
Polymorphisms of the GLI3 gene were associated with weight at birth and 6 months in Chinese cattle (Huang et al., 2013), ELF1 is involved in lipid metabolism (Jin et al., 2009; Chang and Miyamoto, 2006) and regulates adipocyte differentiation (Baek and Baek, 20136). It has also been reported that ELF1 regulates intramuscular fat in cattle (Ramayo-Caldas et al., 2014; Rudra et al., 2012). DIAPH3 is associated with height variation (Metzger et al., 2018) and also down-regulated in the longissimus dorsi muscle in Korean cattle (Lee et al., 2017). Another potential candidate genes identified were CDK6, SENP6, and DECR1. SNP in the promoter region of the CDK6 gene was found to be significantly associated with body length and heart girth in Chinese and Simmental cattle breeds (Liu et al., 2001). DECR1 is identified to be associated with ultrasound backfat in European beef breeds (Angus, Charolais) (Marques et al., 2009), linked to lipid metabolism and linoleic content (Amills et al., 2005), and contains QTL for fatty acid composition in pigs (Clop et al., 2003). For the last few decades, the Brown Hanwoo cattle have been subject to selection for growth and carcass quality traits. In addition to variation in production traits, the study breeds also exhibit distinct coat colors and patterns. More importantly, functional process analyses revealed that genes differentially differentiated between and Chikso and Hanwoo associated with relevant GO terms including, fatty acid beta-oxidation (PPARD, DECR1, HSD17B4, HADHA), adipose tissue development (PPARD, ARID5B, FTO), melanocyte differentiation (MREG, GLI3, OCA2), and melanosome transport (BLOC1S3, MREG) (Table 3).
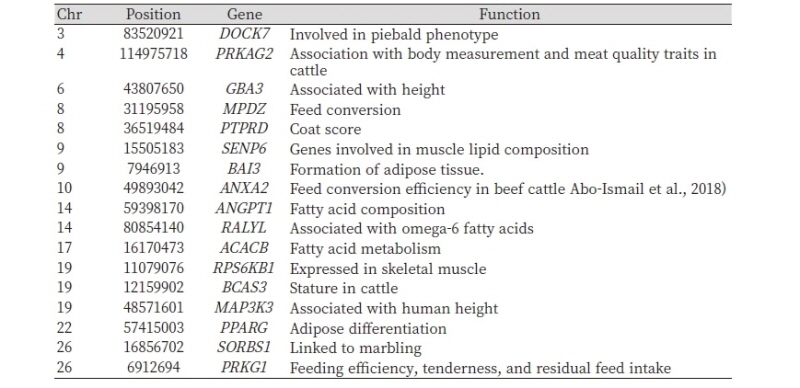
For the Hanwoo-Heugu comparison, we detected genes under positive selection and previously known to affect beef production traits. Three candidate genes (PRKAG2, GBA3, and BCAS3) were shown to be associated with stature (Hawlader et al., 2016; Fu et al., 2014; Jennie et al., 2011). Several of our candidate genes have been demonstrated to affect meat quality and feed efficiency (Table 4). ACACB, PPARG, SENP6, PRKG1, and ANGPT1 have previously been identified to linked to fatty acid metabolism, adipose differentiation, muscle lipid composition, and marbling (Zhang1 et al., 2017; Moisa et al., 2013; Dunner et al., 2013).
The results of the present study may provide some insight into the effect of artificial selection /demographic events in shaping the genetic diversity of Korean Peninsula cattle breeds. The smaller effect effective population sizes obtained for Chikso and Heugu over generations, warrants further strengthening of the ongoing conservation program. Following validation using more samples, SNPs which are identified as breed-specific can be used for product traceability and/or breed assignment. Given the evidence that the brown Hanwoo cattle have been intensively selected for beef traits over the last few decades, the genes identified in this study might be potential growth and carcass traits candidates for follow-up studies and subsequent inclusion in the selection program.
Acknowledgements
This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry(IPET) through “Development of on-site verification system and device for Identifying Hanwoo beef” Project, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA)(318015-3)