Introduction
In recent years, significant progress has been achieved in animal husbandry of the Republic of Belarus, which is not the case in the republic of Iraq where meat is the main purpose for sheep breeds raising this require improving sheep’s meat quality which is controlled by several indicators such as water-holding capacity, fat content and distribution, color, tenderness and texture that are the key parameters affecting meat quality (Miller et al. 1995).
Meat quality is a very important economic traits in meat industry which can be affected by several factors including breed, feeding, fasting, preslaughter handling, stunning, slaughter, and storage condition and has a quantitative characteristic that is controlled by many genes.
One of the major indicators of meat quality is tenderness which is affected and controlled by many factors such as reproduction, protein composition of muscle fibers before and after slaughter, Scientific discoveries in molecular genetics have made a significant contribution to the search for genes and markers that can act as a key player in meat quality especially meat tenderness, one of the genes and marker that control this parameter is the calpastain gene (CAST) as the as the recent genetic studies declare (Stoikova-Grigorova and Stefanova 2018) (Singh et al. 2014). The CAST gene is located on the fifth chromosome of sheep and has a significant effect on the tenderness of meat by inhibiting calpains in the posthumous process (Bhat et al. 2018).
The calpain proteolytic system in the cell is considered to be one of major reasons responsible of post-mortem meat tenderization process (Huff-Lonergan and Lonergan 2005) which is controlled by the ubiquitous calcium-dependent proteases l-calpain and m-calpain that are encoded by The CAPN1 and CAPN2 genes, and its specific inhibitor, calpastatin (CAST), encoded by the CAST gene (Paredi et al. 2012) (Wells and Leloup 2010). The calpain activity provides the diffusion of myofibrillar protein which lead to improvement of tenderness therefore the high levels of calpastatin are associated with decreased proteolysis and increased meat toughness (Kemp et al. 2010) (Kent, J Spencer, and Koohmaraie 2004). and by that the less the cast express the high the tenderness goes especially by the strong genetic correlations between calpastatin activity and tenderness (Casas et al. 2006).
The CAST gene is considered as strong marker that can be used to ameliorate meat tenderness quality, for that it is necessary to identification this gene. Several methods are used for gene identification yet the polymerase chain reaction and restriction fragment length polymorphism (PCR-RFLP) is the most used technique for been fast, specific and direct method that demand few consumables and steps. (Pegg et al. 2016; Sourri et al. 2019; Zhao et al. 2019)
Thus, the goal of our research is to develop a technique that allows studying and identifying allelic variants of the CAST gene in sheep at the molecular genetic level.
Material and method
PCR-RFLP method was used to determine Polymorphism of the CAST gene
Animal samples and PCR-RFLP assay
The study was conducted on 150 animals of three sheep breeds selected from three regions as following: Texel breed from Hvineviehy farm Hrodno region, Ile de France breed from Istern farm Minsk region Belarus and Awassi breed from Agriculture research center from the Baghdad-Iraq, respectively (n = 150), the tissue samples were extracted from 150 lambs and PCR products were digested with MspI enzyme. Quality and quantity of DNA were measured by spectrophotomter method.
The selection of nucleic acids was carried out by the double-cleaning perchlorate method where the concentration of isolated nucleic acids was recorded using an Implen P330 spectrophotometer. Amplification of the CAST gene was performed using the following sequence of primers:
Ovine 1C: 5ґ-TGGGGCCCAATGACGCCATCGATG-3ґ
Ovine 1D: 5ґ-GGTGGAGCAGCACTTCTGATCACC-3ґ
PCR was carried out in a reaction medium where in a total volume of 25 μl which consisted of 50-100 ng of template DNA, 2.5 ul PCR buffer 10X (200 mM (NH4)2SO4, 0.1 Mm Tween 20, 750 mM Tris-HCl (pH 8.8), 2.5 mM MgCl2, 200 μM dNTPs, and 10 pM of each forward and reverse primers and 1 U of Taq DNA polymerase.
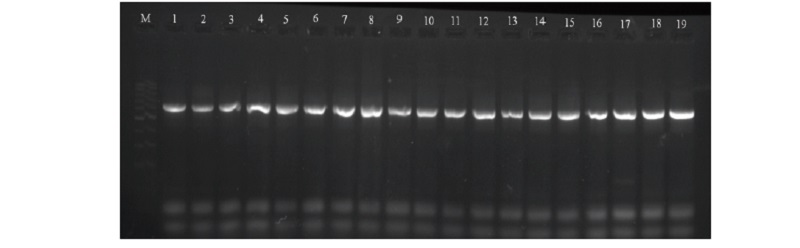
PCR mode for amplification of the CAST gene: “Hot start” started with a primary denaturation for 4 minutes at 95° C, followed by thirty-five cycles denaturation at 94° C for 45 sec, annealing at 62° C for 45 sec, synthesis for 45 sec at 72° С followed by 72°C (7 min) for the final extension. Exon 1C/1D from domain 1 region including the intron of the ovine CAST gene were amplified to a 622 bp fragment where the amplification product was separated on a 2% agarose gel for 50 minutes using a voltage of 110V.
The fragments obtained during amplification were subjected to Msp I restriction enzyme at a temperature of 370 C for 12–16 hours, the obtained results were visualized in a 3% agarose gel at a voltage of 130 V using the gel-documenting system GelDoc XR +, Bio-Rad.
Results and discussion
In this study, we aimed for detecting and identifying a key player gene polymorphism in meat eating parameters with a fast, specific and direct method that doesn't demand a lot of consumables and steps. Moreover, in this method, only one pair of primers was used, which could avoid potential bias caused by amplification efficiency rendered by different primers (Zhao et al. 2019).
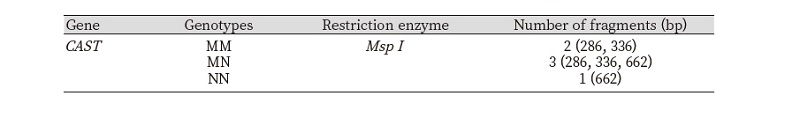
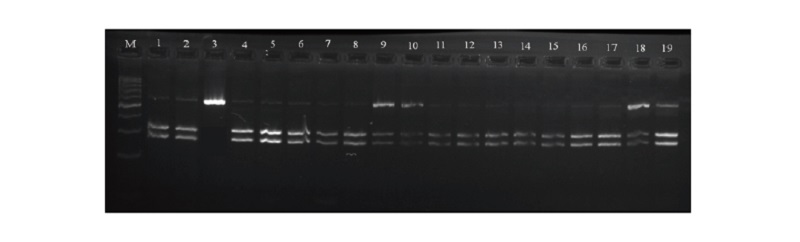
One hundred Fifty sheep were selected from three deferent breeds from three different regions, Texel breed from Hvineviehy farm Hrodno region-Belarus, Ile de France breed from Istern farm Minsk region-Belarus and Awassi breed from Agriculture research center Baghdad-Iraq, respectively (n = 150), to identify the CAST (calpastain) polymorphism gene, using the polymerase chain reaction and restriction fragment length polymorphism PCR-RFLP technique which is known as a rapid yet specific and easy assay (Pegg et al. 2016; Sourri et al. 2019; Zhao et al. 2019) for identification that consist on using just a primers and restriction enzyme where the Msp I was the enzyme selected in our case. The fragments obtained as splitting of amplification products of the CAST gene recognizes three genotypes as it is showed in the table 2, where 336 and 286 bp assigned to the MM genotype, 622 bp assigned to NN genotype and 622,336 and 286 bp for the MN genotype as it is illustrated if Fig.1 and 2.
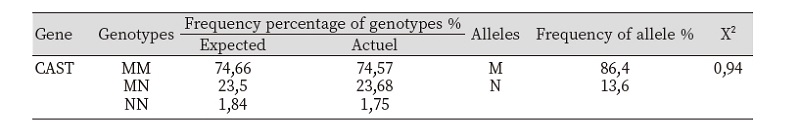
The frequency of genotypes and alleles of CAST genes, is shown in table 2 which exhibit and indicate that in the population of tested sheep of the three breeds, polymorphism was established for the calpastain gene, was represented by two alleles, M and N, where 86.4% of the alleles for this gene are the M alleles, while 13.6% are the N alleles, And by that the genotype MM, was the most frequent one with 74.57 % of the studies sheep, than the MN genotype that present in 23.68%, while the NN genotype was the less often with 1.75%. On the other hand, An earlier studies was done where only two genotypes were observed the CAST MM with 86,4 the CAST MN 13,6 and the CAST NN genotype has not been observed (Gabor, Trakovická, and Miluchová 2013), while only genotype MM was observed in the Bulgarian sheep breed (Bozhilova and Dimitrova 2015).Thus, the more knowing and assessment the genotype of the calpastatin gene (CAST) of sheep are reveal and identified will make it possible to more efficiently use the alleles profile for the studied genes, and by that increasing or decreasing their frequency and by that affecting the gene expression or the acid sequence of a product and ultimately impact on meat quality traits (Romero, Ruvinsky, and Gilad 2012).
the result of this research could be summarized, by saying that polymorphism of the CAST gene was determined using the PCR-RFLP analysis was developed and adapted. When restriction is carried out, three genotypes were identified, CAST MM, CAST MN and CAST NN (preferred), the more this gene genotype polymorphism are reveal and identified the more controlling and improving meat quality is possible, affecting and manipulating the allelic frequency which means gene expression or mutation which might be responsible of maintaining the calpastain -the inhibitor of the ubiquitous calcium-dependent proteases- level leading to the increasing of the proteolytic activity of key proteins within muscle fibers in post-mortem stage which is associated with meat tenderness (Calvo et al. 2014) (Kemp et al. 2010), this gene is also considerate to be an important candidate implicate in the color propriety of meat (Castro et al. 2016)
Conclusion
In conclusion, solving an important economic task and problem of increasing meat eating quality in the cattle breeding filed and meat industry by employing modern molecular genetics became The focus of attention recently, The established work was designed to identified allelic variation of the cast gene using RPC-RFLP method in sheep population of Iraq and Belarus due to importance of the sheep in livestock and meat industry and considering that the DNA markers of productivity will make it possible to significantly increase the genetic potential of speed where three genotypes CAST MM, CAST MN, CAST NN were identified which can useful for improving meat quality especially tenderness. Further researches are conducted on studying the polymorphism of the CAST gene and its relationship with meat productivity.