Abstract
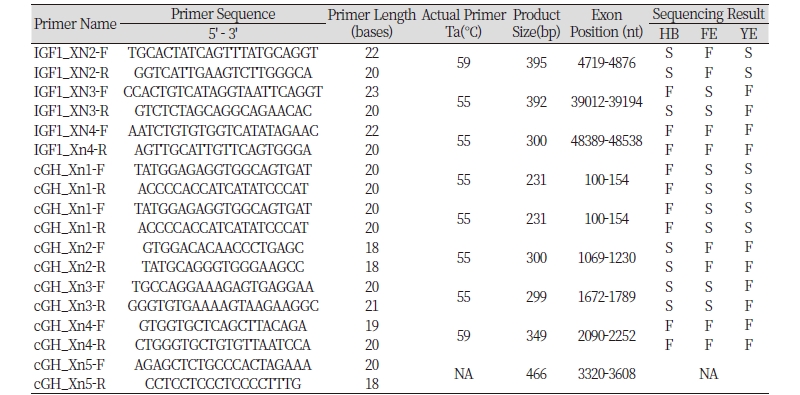
Somatotropic Axis Genes are candidate genes for differential growth determination. Component exons (and 50 bases immediately 5’ and 3’ of each exon) of IGF1 (4 Exons) and cGH (5 Exons), were amplified from DNA of fast (Hubbard Broiler, HB), and slow (Nigerian Local Chickens: Fulani Ecotype, FE and Yoruba Ecotype, YE) growing chickens. Sequenced and aligned against each other and against Genbank published orthologs spanning Aves, Mammals, Pisces and other vertebrates. Phylograms were constructed from aligned sequences. Regulatory motifs within noncoding DNA sequence were identified. Of the exons examined, all but exon 5 of cGH were successfully amplified in all breeds. However, only 33% of IGF1 (HB, exon 2 and 3; FE exon 3; YE exon 2) and 42% of cGH (HB, exon 2 and 3; FE exon 1 and 3; YE exon 1) exons were successfully sequenced. No inter-breed (between HB, YF, and FE) coding sequence polymorphisms were detected. Equally, none was detected in comparisons with published Jungle Fowl sequence (RJF; ENSGALT00000000328 and ENSGALT00000020816, Ensembl.org). Four polymorphisms were detected in the 5’ and 3’ flanking DNA of sequenced exons, including: a cGH 1644 A>G (RJF > HB, FE) single nucleotide polymorphism or SNP; a cGH 1850 G>A (RJF > FE) SNP; a cGH 1860 T>C (RJF >FE) SNP and; an IGF1 4662 – 4673insT insertion polymorphism. Three of these polymorphisms overlapped regulatory motifs. The lack of polymorphisms in aligned sequences suggests these particular Exons are not responsible for the differences in growth rate observed between the breeds.
Figures & Tables