Abstract
Horses are raised for various fields such as horse racing, and it was breed mainly for physical ability. Horses are suitable models for exercise research. In skeletal muscle of horses, many genes are expressed after exercise. Previous studies, we found differentially expressed genes (DEGs) related to exercise in horses. The transcription factor (TF) binding site of the 5'-regulatory region of the B3GNT5 was predicted through PROMO. In addition, the genotypic frequency of nsSNP (rs69214296) and the Hardy-Weinberg equilibrium test were performed using 98 Thoroughbred race horses. We conducted a study to identification gene function using gene expression analysis and bioinformatic analysis for horse UDP-GlcNAc:BetaGal Beta-1,3-N-Acetylglucosaminyl transferase 5 (B3GNT5). The expression of B3GNT5 was increased about 3.37 times after exercise compared with before exercise. Six transcription factors were predicted at the upstream 600 bp of the B3GNT5 gene. In addition, the genotype frequencies of non-synonymous SNPs (nsSNPs) were analyzed in B3GNT5 to confirm the significance between the predicted and observed genotypes. We identified the characteristics of B3GNT5 gene in horses through this study. Further analysis is needed to investigate the impact of the B3GNT5 SNP on racehorses.
Figures & Tables
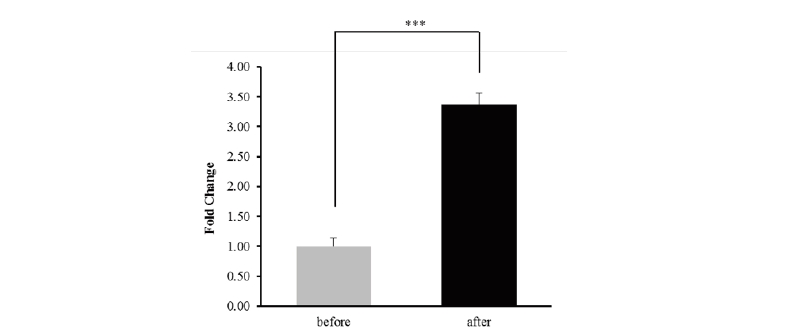
Figure 1. Gene expression of B3GNT5 before and exercise as decided via whole transcriptome analysis. *** p<0.001


