Abstract
Growth-related traits (daily gain, back fat thickness, and carcass weight et al.,) are complex and economically important in the swine industry. The purpose of this study was to confirm quantitative trait loci (QTL) and the associated positional candidate genes affecting daily gain in pigs. A genome-wide association study (GWAS) was performed using the porcine SNP 60K bead chip. The data used in the study included 1,755 purebred Yorkshire pigs. All experimental animals were genotyped with 44,985 SNPs located throughout the pig autosomes. We identified association between a SNP markers on chromosome 15 (24.8-25.5 Mb). Three positional candidate genes are located on SSC15, STEAP3, C15H2orf76 and TEX51. This genes may provide in the selective breeding program after validating its effect on other populations.
Figures & Tables
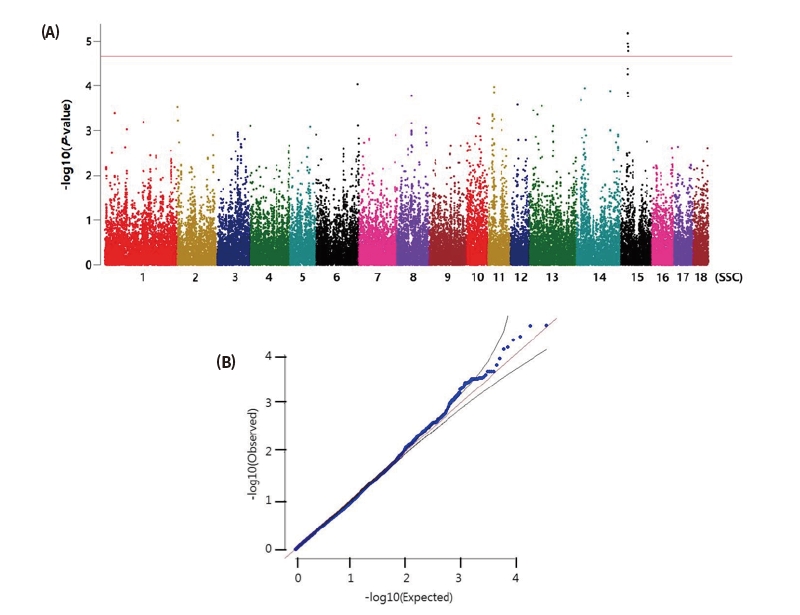
Figure 1. Manhattan plot(P-value) and Quantile-Quantile plot of genome wide association study of daily gain. SNPs locate their position on each chromosome against on the x-axis and the association signal on the y-axis(shown as -log10 of the P-value). (A) Association between daily gain and 44,985 mapped SNP markers in 18 pig autosomes using additive model. (B) Quantile-Quantile plots of SNP markers.


