Introduction
The ability to edit genomic sequences precisely, based on programmable DNA nucleases, such as zinc finger nucleases (ZFNs), transcription activator-like endonucleases (TALENs) and the clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein 9 (Cas9), has revolutionized various fields of genome engineering in living organisms (Lee et al., 2015; Kim, 2016; Lee et al., 2017). These genome editing systems hold great promise for furthering our understanding of the function of a gene in biological research, and have practical applications in the fields of animal biotechnology, biomedicine and gene therapy (Park et al., 2014; Lee et al., 2017; Lim, 2017; Winblad and Lanner, 2017).
Recent advances, particularly the CRISPR-Cas9 system, have led to the creation of some of the most powerful tools for genome editing to date by enhancing the DNA mutation rate through creation of a double-stranded DNA break (DSB) at a specific locus in the genome and highly efficient genetic modification in a targeted manner (Jinek et al., 2012; Kim, 2016). Concurrently, significant efforts have been made to unveil the underlying molecular mechanism of this process (Deltcheva et al., 2011; Gasiunas et al., 2012), and make it easy to transfer into eukaryotic cells (Cong et al., 2013; Mali et al., 2013). Therefore, it has been successfully adopted for precise genome editing in diverse organisms, including animals, plants and humans (Lee et al., 2017; Ma et al., 2017; Niu et al., 2017; Ruan et al., 2017). Ensuing from the revolutionary CRISPR-Cas9 system, a scientist has coined the term “the CRISPR Zoo” in the specialized field of animal biotechnology, because current genome editing technology is highly improved and easily utilized (Reardon, 2016).
Most recently, a novel base editing system that enables the programmable conversion of one base pair into another and local sequence diversification without double-stranded DNA cleavage was developed (Komor et al., 2016; Nishida et al., 2016; Hess et al., 2017). In most known genetic disorders, the correction of a point mutation enables us to address the underlying cause of the disorder (Landrum et al., 2016). However, Cas9-mediated genome editing via the delivery of a homology- directed repair (HDR) donor template is very low in efficiency for the correction of a point mutation. Although Cas9-mediated genome editing has been optimized for single-nucleotide editing efficiencies, reaching 60% or more in cell culture systems (Richardson et al., 2016), this strategy generally depends on inducing a DSB. A natively generated DSB triggers small insertions or deletions (indels) that are an unintended by-product, which can have major negative functional consequences (Hess et al., 2017). Therefore, precise base conversion based on a base editing system provides a valuable tool for genetic modification regarding highly targeted single-base changes and will draw much attention from a large number of scientists.
In this review, we introduce recent advances in genome editing technologies based on targeted DNA nucleases and a base editing system, and highlight their applications to various fields of genome engineering.
Development of programmable genome editing tools
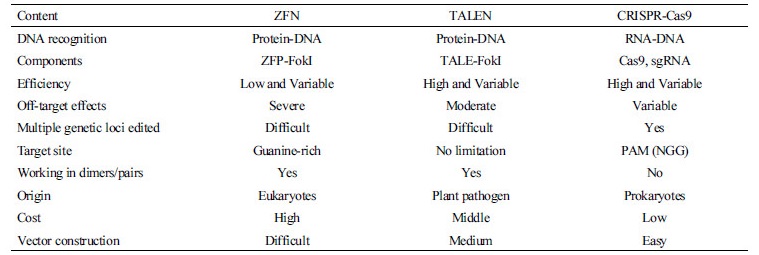
During the past decade, programmable genome editing tools such as ZFN and TALEN have made genome editing possible. Briefly, the first emergence of a programmable genome editing tool was ZFN, in which specific DNA-binding zinc finger proteins are fused with a nuclease domain such as the FokI endonuclease (Kim et al., 1996; Carbery et al., 2010). As shown in Figure 1, pairs of ZFNs specifically bind to the target region and enable the induction of DNA cleavage via FokI dimerization, resulting in the generation of DSBs. The pairing of ZFN motifs with three nucleotides depends on the amino acid composition in the genomic context (Smith et al., 2000). Furthermore, ZFNs enable efficient gene targeting in specific genomic loci compared to homologous recombination (Porteus and Carroll, 2005). This ZFN technology has been widely applied to the introduction of genetic modifications in diverse organisms (Urnov et al., 2010). However, there are limitations regarding ZFN delivery into cells, including severe off-target effects and toxicity.
The second type of programmable genome editing tool is TALEN, in which the TAL effector of a DNA-binding domain derived from the pathogenic bacteria Xanthomonas is fused with a DNA cleavage nuclease domain (the same FokI endonuclease used in ZFN) (Boch et al., 2009). TALEN also specifically recognizes target sequences in the genome and enables the induction of DNA cleavage, resulting in the generation of DSBs similar to sequential events in ZFN (Figure 1). The TAL effector, which is composed of the DNA-binding domain, contains 33–34 conserved amino acid sequences with a variable region at amino acid positions 12 and 13 called the repeat variable di-residue (RVD). The DNA specificity of a TAL effector depends on the amino acids of the RVD to recognize different nucleotides (HD binds to C, NG binds to T, NI binds to A, and NN binds to A or G) (Boch et al., 2009). Moreover, TALEN is a more useful and flexible tool (compared to ZFN) for editing genome sequences in the target site as a designed nuclease because each TAL effector, which contains one repeat domain, can bind to one nucleotide (Miller et al., 2011). Therefore, TAL effectors have been widely utilized to modify a specific genomic locus, because TAL effectors can combine with activator domains such as transcriptional activators and endonucleases (Mussolino and Cathomen, 2012). Consistent with the development of the TALEN assembly kit using golden gate shuffling, customized TALEN can be easily synthesized and used to make precise genetic modifications (Engler et al., 2009; Cermak et al., 2011; Sanjana et al., 2012).
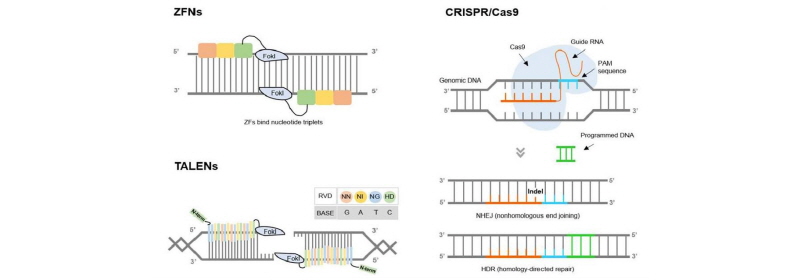
Figure 1.
Development and principles of genome editing technologies: zinc finger nucleases (ZFNs), transcription activator-like endonucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR)- CRISPR-associated protein 9 (Cas9). Recent advances, particularly the CRISPR-Cas9 system, have become some of the most powerful tools for genome editing to date by enhancing the DNA mutation rate through the creation of a double-stranded DNA break (DSB) at a specific locus in the genome and highly efficient genetic modification in a targeted manner. Today, almost every scientist in the world can conduct genome editing experiments to create new developments and opportunities in any animal species.
More recently, the CRISPR-Cas9 system, as a simple RNA-guided platform, has been considered the most revolutionary tool and was developed to achieve highly efficient and specific genome editing in a targeted manner. The components of CRISPR and Cas9 are derived from prokaryotic DNA, which plays an important role in acquired resistance against the invasion of plasmid DNA and phage into bacteria. These bacteria have clustered repeats called CRISPRs, which are identical to viral genomes, to defend themselves. When a virus enters a cell, a CRISPR binds to the viral RNA to disrupt it with the Cas9 protein (Barrangou et al., 2007). The CRISPR-Cas9 system also specifically recognizes and sequentially drives DNA cleavage at the target site, resulting in the generation of DSBs (similar to ZFN and TALEN). However, unlike ZFN and TALEN, CRISPR-Cas9 does not require paired units to induce DNA DSBs at the target site. The CRISPR-Cas9 system is a type II CRISPR system that includes the Cas protein, CRISPR RNA (crRNA), trans-activating CRISPR RNA (tracrRNA), and a protospacer adjacent motif (PAM) sequence for genome editing (Jinek et al., 2012). The PAM sequence is generally located upstream of the crRNA-binding region and guides the Cas9 protein for target recognition with a “seed” sequence within the crRNA. In detail, CRISPR-Cas9 protein-RNA complexes specifically recognize a target DNA sequence through base pairing with a guide RNA, and natively induce DNA cleavage to create a DSB at the targeted locus by the guide RNA (Figure 1). Subsequently, these DSBs are involved in cellular DNA repair processes, resulting, mostly, in random indels at the site of DNA cleavage via non-homologous end joining (NHEJ) or replacement of a homologous DNA template in the DNA surrounding the cleavage site via HDR. Moreover, the CRISPR-Cas9 system has many advantages for sequence-specific genome editing, including gene knockout, gene knockin and site-specific sequence mutagenesis and corrections, making it extremely easy to synthesize crRNA and tracrRNA and comparatively simple for the construction of thousands of customized CRISPR-Cas9 systems depending on the targeted genes. In addition, the CRISPR-Cas9 system is a fast and cost-effective system for modifying the genomes of various organisms because of its simple and practical ease of use and robust cutting activity compared with ZFN and TALEN DNA modification technologies that rely on DNA-binding specificity, which are developed through expensive and time-consuming processes (Cho et al., 2013; Hwang et al., 2013; Wang et al., 2013; Sung et al., 2014). A comparison of ZFN, TALEN and CRISPR-Cas9 systems is summarized in Table 1.
Based on the comparison of ZFN, TALEN and CRISPR-Cas9 systems, the CRISPR-Cas9 system has become the most powerful tool for precise genome editing in a targeted manner and is successfully utilized for precise genome editing in diverse organisms, including animals, plants and humans (Lee et al., 2017; Niu et al., 2017; Shimatani et al., 2017; Winblad and Lanner, 2017).
Base editing system
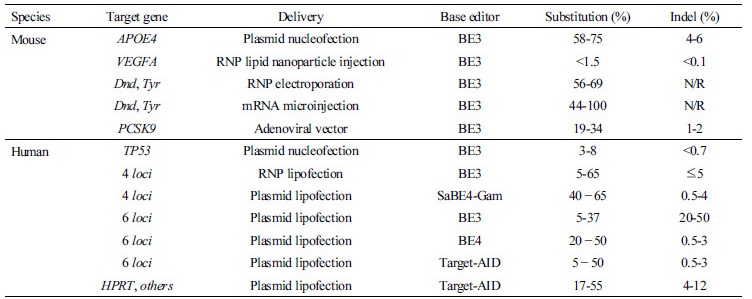
Most genetic diseases arise from various types of genetic or environmental mutations, such as indels, substitutions and frameshifts in the genome caused by single-nucleotide substitutions or point mutations (Men et al., 2017). However, previous strategies used to correct point mutations are difficult to apply effectively to genetic diseases. Although genome editing mediated by CRISPR-Cas9 was optimized for single-nucleotide editing efficiencies, reaching up to 60% in cultured cells (Richardson et al., 2016), this strategy generally depends on induced indels resulting from the cellular response to DSBs (Hess et al., 2017). Therefore, to minimize DNA damage during genome editing and to achieve point mutation corrections without the use of an HDR donor template, a programmable base editing system to induce precise base conversion efficiently was recently developed and applied to various species. A base editing system was designed using the APOBEC (apolipoprotein B mRNA editing enzyme) 1-4 or AID (activation-induced deaminase), a group of cytidine deaminases that insert mutations in DNA/RNA as a result of their ability to deaminate cytidine to uridine (Conticello, 2008). As shown in Figure 2, these deaminases are fused to CRISPR-Cas9 to substitute a C at a target site with T (or G→A) without double-stranded DNA cleavage (Mali et al., 2013; Ran et al., 2013; Komor et al., 2016; Ma et al., 2016). Base editors with improved conversion efficiency have been developed continually, and up to the fourth generation of base editors, which include BE1, BE2, BE3, and BE4, have been reported. In detail, Komor et al. reported a first-generation base editor (BE1) using the rat APOBEC (rAPOBEC) cytidine deaminase enzyme fused to the N-terminus of catalytically dead Cas9 (dCas9) by a 16-residue peptide XTEN linker (Komor et al., 2016). Of the four enzymes hAID, hAPOBEC3G, rAPOBEC1, and pmCDA1, rAPOBEC1 exhibit the highest deaminase activity; connecting rAPOBEC1 to the N-terminus, not the C-terminus, of dCas9 preserves its deaminase activity. Therefore, rAPOBEC1-dCas9 has been fused to linkers of different lengths and compositions, and BE1 converted the DNA base cytosine (C) to uracil (U), which has the base-pairing properties of thymine (T), with an activity window of approximately five nucleotides, at target positions 4 to 8. The average base editing efficiency of BE1 in vitro is 44%, but its efficiency decreases by 5- to 36-fold to 0.8-7.7% in human cells (Komor et al., 2016). Base excision repair, which removes uracil (U) from the DNA, was believed to be the cause of decreased intracellular base editing efficiency, and the second generation of base editors, which fuses UGI to the C-terminus of BE1 (BE2, APOBEC- XTEN-dCas9-UGI), was created to increase the low editing efficiency of BE1 in targeted cells. As a result, the editing efficiency of BE2 in human cells, such as human embryonic kidney 293 cells, increased, on average, 3-fold compared to BE1, resulting in improved gene conversion efficiency of up to 20%. BE1 and BE2 have the advantage that indel formation rates were less than 0.1% because the DNA is not directly cleaved (Kunz et al., 2009; Komor et al., 2016). To increase the editing efficiency to more than 50%, a third generation of base editors (BE3, APOBECXTEN–dCas9 (A840H)–UGI) that nicks an unmodified DNA strand was generated. In addition, BE3 retains the Asp10Ala mutation at Cas9, which prevents double-stranded DNA cleavage. As a result, the editing efficiency of BE3 in human cells increased, on average, 3- to 6-fold compared to BE2, resulting in up to 37% efficiency. The indel formation rate of BE3 was 1.1%. When Komor et al. compared HDR and BE3 for two disease-relevant mutations in APOE4 and p53, Cas9-mediated HDR showed a very low efficiency (0.1-0.3%); however, BE3 efficiency was 58-75% in APOE4 and 3.3-7.6% in p53 (Komor et al., 2016). Subsequently, fourth generation base editors (the Streptococcus pyogenes Cas9-derived base editor BE4 and the Staphylococcus aureus Cas9-derived BE4 and BE4gam) were developed to improve the editing efficiency of C:G to T:A to approximately 50% efficiency, halving the frequency of undesired by-products compared to BE3 (Komor et al., 2017). In addition, the bacteriophage Mu protein Gam was fused to the N-terminus of BE4 to reduce the indel frequency. Gam protein binds to the ends of DSBs and protects them from degradation during the base editing process. Indeed, adding Gam to BE4 or SaBE4 decreased the indel frequency by 1.5- to 2-fold (Komor et al., 2017). Moreover, for the purpose of broadening the applicable scope of base editing, a technique that can reverse A∙T to G∙C was recently developed (Gaudelli et al., 2017). A base editing system has already been successfully applied in vitro and in vivo for precise genetic modification of interesting genes in humans and in mice (Table 2). For the specialized purpose of therapeutic applications, base editing systems have been applied in a wide range of mammalian cell culture models and successfully demonstrated in vivo by adenoviral vector delivery or ribonucleoprotein (RNP) delivery into mouse tissues. For the purpose of agricultural traits engineering, base editing systems have also been successfully adopted in diverse plants, such as tomato, wheat, maize, and rice (Ding et al., 2014; Nishida et al., 2016; Komor et al., 2016; Chadwick et al., 2017; Kim et al., 2017; Liang et al., 2017; Lu and Zhu, 2017; Rees et al., 2017; Shimatani et al., 2017; Zong et al., 2017). Most recently, CRISPR-STOP and iSTOP systems, which induce stop codons by CRISPR-mediated base editing, were used for the disruption of eukaryotic genes, with 97-99% of genes in eight eukaryotic species being precisely targeted (Billon et al., 2017). Utilizing this system, a stop codon was generated by converting C to T at the target site of the Dmd or Tyr genes using mRNA or RNP in mouse embryos. As a result, F0 mice showed a nonsense mutation with an efficiency of 44-57%, and allelic frequencies of 100% were observed (Kim et al., 2017). In the near future, the base editing system will provide a valuable tool for precise genome editing with regard to highly targeted single-base changes, and could be widely used for specialized purposes regarding the production of model animals associated with amino acid substitution in any species of interest (Figure 2).
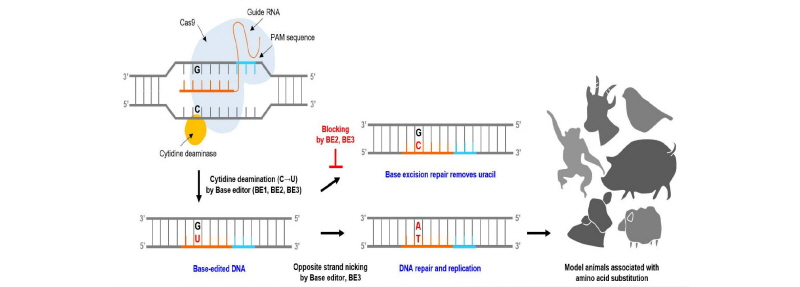
Figure 2.
Development and application of the base editing system. A single base editing system is generated by fusion of cytidine deaminase to the CRISPR-Cas9 system. Target C base is deaminized with U by three base editors (BE1, BE2 and BE3). Base excision repair (BER) reduces the intracellular base editing efficiency by removing uracil. BER can be blocked by BE2 and BE3 containing uracil DNA glycosylase inhibitor (UGI). BE3 also allows DNA repair and replication through opposite strand nicking. The base editing system provides a valuable tool for precise genome editing with regard to highly targeted single-base changes and can be widely used for specialized purposes in the production of model animals associated with amino acid substitution in any species of interest.
Genetic engineering and applications
As mentioned above, the CRISPR-Cas9 system has many advantages in precise genome editing due to its robust, efficient and cost-effective manner. Furthermore, the CRISPR-Cas9 system has been successfully established in a number of living organisms. Here, we highlight successful research achievements and potential uses with programmable genome editing tools in various fields of genome engineering.
Chicken
Avian species have been historically used as an animal model to study early embryogenesis and organogenesis in developmental biology (Stern, 2005), as well as in avian biotechnology (Han, 2009). However, it has been difficult to investigate the function of a gene in aves because of their unique developmental processes and the absence of a precise genome editing system. Consistent with the development of programmable genome editing tools, efficient gene targeting approaches in chickens have been successfully adopted. In 2013, the immunoglobulin gene knockout chicken was first produced via homologous recombination in chicken primordial germ cells (PGCs) (Schusser et al., 2013). However, the total germline transmission rate of targeted PGCs is approximately 0.1%, indicating that homologous recombination events occur at a very low frequency. Next, a targeted ovalbumin knockout chicken was generated by TALEN in 2014 (Park et al., 2014). A gene knockout system mediated by TALEN exhibits higher germline transmission efficiency in the mutant progeny compared to the conventional homologous recombination-mediated gene knockout system, demonstrating that TALEN- induced NHEJ occurs much more frequently than homologous recombination in eukaryotic cells (Kim and Kim, 2014). More recently, an ovomucoid (OVM) gene-targeted chicken mediated by the CRISPR-Cas9 system was successfully produced (Oishi et al., 2016). Concurrently, Dimitrov et al. successfully produced a precise genome-edited chicken mediated by CRISPR-Cas9 via HDR insertion of an additional loxP site into the variable region segment (VH) of a loxP in a previously produced gene targeting chicken resulting in successful insertion of the joining gene segment (JH) into the immunoglobulin heavy chain (IgH) locus by homologous recombination (Schusser et al., 2013; Dimitrov et al., 2016). Moreover, Tayler et al. successfully produced a CVH gene-targeted chicken via the TALEN-mediated HDR system, which replaced green fluorescent protein (GFP) transgene integration into the CVH locus on the Z chromosome (Taylor et al., 2017). Furthermore, Cooper et al. recently proposed an alternative method for efficient genome editing called sperm transfection-assisted gene editing, which is based on direct delivery of a CRISPR gRNA and Cas9 mRNA mixture into spermatozoa in chickens (Cooper et al., 2017). Together, programmable genome editing tools have been successfully utilized to achieve precise genome editing for various purposes in chickens.
Cattle
Bovine genome modification has been mainly employed to achieve transgenesis in agricultural and biomedical fields for practical applications. In the past decade, the production of genetically modified cattle has been advanced by technological developments. Due to the lack of embryonic stem cells (ESCs) in cattle, the first successful transgenic cattle were produced by pronuclear injection (PNI); however, the efficiency was rather low and this system was unable to conduct site-specific genetic modification (Hammer et al., 1985). To overcome the extremely low efficiency of transgenic progeny by the PNI system, sperm-mediated gene transfer and viral vector-mediated gene delivery were explored. However, these approaches still yielded low reproducibility regarding the production of transgenic animals (Bachiller et al., 1991; Kubisch et al., 1997; Hofmann et al., 2003). Later, somatic cell nuclear transfer (SCNT) or chromatin transfer was developed to improve genetic modification, resulting in a remarkably higher targeting efficiency (Cibelli et al., 1998; Hofmann et al., 2003). However, homologous recombinant (HR) activity is very low because the proliferative capacity of bovine somatic cells is limited in vitro (Sedivy and Sharp, 1989). Despite these limitations, a few research groups have reported that HR-mediated site-specific genetic modification could successfully produce knockout and knockin animals via cattle somatic cells (Kuroiwa et al., 2004; Sendai et al., 2006; Richt et al., 2007; Robl et al., 2007; Wang et al., 2013; Matsushita et al., 2014). With the sequential development of designer nucleases such as ZFN, TALEN and CRISPR-Cas9 systems, significant advancements in cattle have been achieved. Since the myostatin (MSTN) gene regulates skeletal muscle mass in mice, inhibition of the MSTN gene has been focused on improving meat production in cattle. In 2014, Luo et al. reported that the generation of MSTN biallelic mutations using ZFN consisted of a 6-nucleotide deletion in one of the alleles and a 117-nucleotide deletion and a 9-nucleotide insertion in the other allele in cattle (Luo et al., 2014). Another example using genome editing mediated by ZFN involved targeting the human lysozyme gene knockin β-casein locus, which enhanced mastitis resistance in dairy cattle (Liu et al., 2014). Compared to previous attempts in other livestock species, a high level of human lysozyme was expressed in the milk. In addition, ZFN-mediated targeting by inserting the lysostaphin gene into the β-casein locus produced a high level of human lysozyme in the cattle mammary gland (Liu et al., 2013). However, the ZFN system is a time-consuming process because of its design and need for validation. Thus, TALEN began to be used for genome modification because it was easily programmed to target and reduce off-target effects. In 2013, Tan et al. reported that a TALEN-mediated system had been developed to specifically target the PC allele for hornlessness in horned dairy cattle (Tan et al., 2013). It was also employed to replace the bovine serum albumin gene with human serum albumin (HSA) to apply a biopharming source of recombinant HSA (Moghaddassi et al., 2014) and to disrupt MSTN gene expression with a high mutation rate (Xu et al., 2013). In 2014, genome editing mediated by CRISPR-Cas9 in bovine pluripotent cells and embryos was first reported (Heo et al., 2015). These results showed that knockin mediated by CRISPR-Cas9 induced highly efficient editing of the bovine NANOG locus in bovine induced pluripotent stem cells as an alternative to pluripotent stem cells for producing genome-edited animals. Recently, a single Cas9 nickase induced the insertion of the natural resistance-associated macrophage protein-1 (NRAMP1) gene via the SCNT system to obtain increased resistance to tuberculosis (Gao et al., 2017). Together, the efficient production of genetically engineered cattle based on programmable genome editing systems can provide a high-quantity production system for therapeutic proteins and improve disease resistance.
Pig
Genetically engineered swine have been used in the production of economic traits and in studies on biofunctional medicine. Over the last century, genetic change was produced by traditional breeding that selectively developed particular phenotypic traits. Moreover, for more than two decades since the development of a variety of techniques to create gene-targeted pigs, efficient genetic alterations have been successfully employed to modify endogenous genes in zygotes or somatic cells. In swine, the first genetic modification was driven by injection of DNA into a pronucleus of a fertilized egg (Hammer et al., 1985; Vize et al., 1988; Petters et al., 1997). However, this system has several limitations, such as lower efficiency of the genetically modified founder, random genome integration or multiple copy insertion (Bishop and Smith, 1989; Wall and Seidel, 1992; Lavitrano et al., 1997; Cabot et al., 2001). To overcome these limitations, genetic modification combined with the SCNT system has been used to successfully produce transgenic pigs (Park et al., 2001). In addition, an HR system with antibiotic selection and SCNT was introduced to create gene knockout and knockin models (Dai et al., 2002; Lai et al., 2002; Rogers et al., 2008), although the efficiency remains low. However, antibiotic selection-completed HR in pigs is directly applicable to biomedical models (Lorson et al., 2011; Flisikowska et al., 2012). The development of programmable genome editing tools has improved the efficiency of gene targeting in swine. A knockout mutation of an enhanced GFP (eGFP) transgene using ZFN combined with SCNT technology has been developed, first reporting on ZFN-based gene modification in swine (Whyte et al., 2011). The ZFN system has been successfully utilized to achieve selective knockout of endogenous swine genes in the genome that are important for agricultural and biomedical applications. In 2013, Lillico et al. demonstrated that both TALEN and ZFN injected directly into pig zygotes produced live genome-edited pigs (Lillico et al., 2013). Double knockout targeting the enzyme α1,3-galactosyl transferase (GGTA1) and the cytidine monophoshated-N-acetylneuraminic acid hydroxylase (CMAH) gene, which are known as xenoantigens, was performed using the ZFN-mediated system for clinical xenotransplantation to reduce the humoral barrier (Lutz et al., 2013). In addition, ZFN-mediated genome editing is able to produce cloned pigs carrying a biallelic knockout of the porcine α1,3-galactosyltransferase (GGTA1) gene, opening a unique avenue toward the creation of gene knockout pigs (Hauschild et al., 2011). In 2013, Xin et al. described TALEN-mediated biallelic knockout pigs with a higher efficiency of GGTA1 mutant progeny (Xin et al., 2013), and Tan et al. reported on targeting deleted in azoospermia-like (DAZL) and adenomatous polyposis coli (APC) genes to produce a model animal to study infertility and colon cancer (Tan et al., 2013). Compared to ZFN and TALEN, the CRISPR-Cas9 system offers higher targeting efficiency and ease of construction. In 2014, Hai et al. demonstrated that mutation of the von Willebrand factor (vWF) gene, which is involved in von Willebrand disease, was produced by zygotic injection of the CRISPR-Cas9 system with the high efficiency of multiplexed genome editing in pigs (Hai et al., 2014). Whitworth et al. reported that direct injection of the CRISPR-Cas9 system into zygotes to target eGFP, CD163 and CD1D efficiently generated genetically modified pigs (Whitworth et al., 2014). In 2015, Zhou et al. firstly targeted the TYR gene exon 1 region, which important for melanin biosynthetic pathway and subsequently performed double knockout of PARK2 and PINK1, which are involved in an autosomal recessive Parkinson’s disease (Zhou et al., 2015). Moreover, MSTN gene editing mediated by the CRISPR-Cas9 system has been reported with a high efficiency in genetically modified pigs (Wang et al., 2015). More recently, Tanihara et al. established a simple method for the electroporation of Cas9 protein in pig zygotes to facilitate genetic modification (Tanihara et al., 2016), and a Chinese group reported that they created healthy pigs with much less body fat by targeting uncoupling protein 1 (UCP1), which regulates adiposity, using the CRISPR-Cas9 system (Zheng et al., 2017). Collectively, genome editing technology has been successfully applied for precise genome editing in swine.
Sheep
Traditional gene targeting techniques in ESCs have been limited to livestock such as swine, cattle and sheep. Additionally, the low efficiency of gene targeting in somatic cells makes it difficult to modify the genome. Thus, in sheep, a genome editing system based on targeted DNA nucleases has mainly been used to generate transgenic animals. In 2004, Zhang et al. reported on MSTN knockout targeting exon 1 to disrupt the expression of the MSTN gene (Zhang et al., 2014). By direct injection of TALEN mRNA into zygotes, they successfully produced MSTN knockout sheep (Proudfoot et al., 2015). In addition, sheep MSTN gene knockout via the CRISPR-Cas9 system was reported (Hongbing et al., 2014). In 2017, the CRISPR-Cas9 system was used to induce dysfunction in the fibroblast growth factor 5 (FGF5) gene as a dominant inhibitor of the length of the anagen phase of the hair cycle, which is an important source of fiber production, resulting in three types of deletions in transgenic sheep progeny (Hu et al., 2017). Collectively, the CRISPR-Cas9 system has been successfully utilized for genetic improvements in sheep breeding and development of the wool industry.
Goat
Genetically engineered goats are important for producing therapeutic proteins and for providing a biomedical model for humans, to contribute to the fields of agriculture and human medicine. A therapeutic protein (an anticoagulant) from transgenic goats was approved by the US Food and Drug Administration in 2009 (Kling, 2009). However, HR gene targeting has several limitations with regard to producing transgenic animals, such as cost and time. Thus, a genome editing system based on targeted DNA nucleases has revolutionized the production of transgenic goats. Xiong et al. reported that ZFN was employed to knockout the β-lactoglobulin (BLG) gene, a dominant allergen in goat milk (Xiong et al., 2013). The TALEN-mediated system was also used to knockout the BLG gene to reduce allergens and to create a knockin of human lactoferrin (hLF) for practical use as a bioreactor (Cui et al., 2015). Furthermore, targeting exon 1 of the BLG locus has been efficiently applied (Zhu et al., 2016). Goat MSTN gene editing has also been examined using the TALEN-mediated system (Yu et al., 2016). In 2014, the CRISPR-Cas9 system was used to induce monoallelic and biallelic gene knockout in goat primary fibroblasts with an efficiency of 9-70% (Ni et al., 2014). The successful generation of genetically modified goats targeting MSTN and FGF5 via the CRISPR-Cas9 system resulted in targeting efficiencies of 15% and 21%, respectively (Wang et al., 2015). In addition, FGF5-disrupted goats exhibited increased secondary hair follicles in the skin and longer cashmere length relative to wild-type goats (Wang et al., 2016). More recently, co-injection of Cas9 mRNA and small guide RNAs into goat embryos were used to create a BLG knockout goat with improved targeting efficiency (Zhou et al., 2017). Thus, genome editing technologies have been successfully adopted for precise genome editing, for practical applications regarding biomedicine and bioreactors in goats.
Other species
Non-human primates are used as essential model animals because their anatomic, physiologic, developmental, social behavior and genetic characters are highly similar to those in humans. In the field of medical research, they have been widely used for human disease research, and genome editing technologies have been applied to primates. Since fluorescent transgenic monkeys were first produced by retroviral gene transfer into mature oocytes (Chan et al., 2001), genome-edited primates have been reported using programmable genome editing tools such as ZFN, TALEN and CRISPR-Cas9 (Niu et al., 2017). In 2015, gene targeting by ZFN at the albumin locus in the liver of non-human primates suggested therapeutic potential for treating Hemophilia B (Wechsler et al., 2015). Furthermore, TALEN-edited MECP2-mutant cynomolgus monkeys were produced as a valuable model animal for studying Rett syndrome, a neurodevelopmental disorder, and phenotypes that included brain developmental and complex behavioral abnormalities were observed in transgenic monkeys (Chen et al., 2017). Although studies on transgenic monkeys are relatively rare compared to other experimental animals, the potential of primates will greatly contribute to human disease research.
Summary
Historically, targeted genome modification has been challenging. Recently, the advent of genome editing technologies, particularly the CRISPR-Cas9 system, has opened a new era in basic science, animal biotechnology, biomedicine, breeding, and therapeutic potential. The CRISPR-Cas9 system has many advantages due to its robust, efficient and cost-effective operation, and provides a valuable tool for precise genome editing. Furthermore, CRISPR-Cas9 and base editing systems have been successfully established in a number of model organisms, as well as in humans. Although there are still concerns regarding the off-target effects, ethics and genetically modified organisms in each field, today, scientists all over the world are able to conduct genome editing experiments to create new possibilities and opportunities from diverse species of interest. Finally, we are certain, in the foreseeable future, that programmable genome editing tools will show tremendous potential in various fields of genetic engineering.