Abstract
We have investigated the genetic diversity and structure using several DNA markers, such as mtDNA, Sry gene in Y chromosome and autosomal SNPs for Japanese Wagyu and native cattle in the North Eastern Asia. Most of Japanese cattle mtDNA belonged to Bos taurus mtDNA haplogroups T1, T2, T3 and T4. All Japanese populations included Asian unique mtDNA haplogroup T4 with high frequencies (0.43 – 0.81). The predominant T4 haplogroup may be a result of founder effect when ancestral cattle were introduced to Japan Islands at the immigration. We found that Mongolian cattle have Bos indicus mtDNA with the genetic frequency of 0.20, but no Bos indicus Y chromosome. No Bos indicus mtDNA in both Japanese and Korean samples suggested that the introgression may be a secondary phenomenon, with the earlier cattle in the region being purely Bos taurus. We analyzed 117 SNPs to assess the genetic diversity, structure and relationships of 16 Eurasian cattle populations. The results could sufficiently explain the genetic construction of Asian cattle populations. These results reflected to the geographical and historical background in each population. The genetic information would contribute to understanding origin of North Asian native cattle, in addition to the origin of Japanese Wagyu cattle.
Figures & Tables
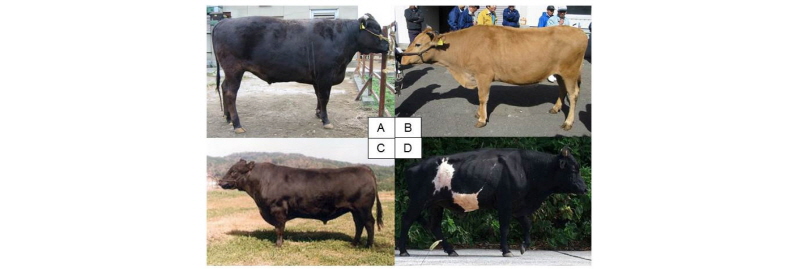
Japanese Wagyu breeds and native cattle. A: Japanese Black cattle. B: Japanese Brown cattle (Tosa strain). C: Japanese Polled. D: Kuchinoshima feral cattle (Photo by Dr. Shimogiri T in Kagoshima University).


