Introduction
The Wagyu generally refers to four Japanese native breeds bred in Japan, but nowadays the famous brand name Wagyu includes not only Japanese native cattle produced in Japan, but also animals or even crossbred Japanese native cattle produced in foreign countries such as Australia or the United States. The Japanese Wagyu, especially Japanese Black cattle, produces high quality beef, which is due to high marbling in muscle. Since Japanese Black cattle have already been improved in terms of beef marbling in Japan, Japanese beef societies are now challenging to improve and look for additional traits on beef quality. Japanese Wagyu has long time history of the breeding and the improvement. In order to understand the genetic background, therefore, we have investigated the genetic diversity and structure using several DNA markers for Japanese Wagyu and also native cattle in the North Eastern Asia. Here, I would like to describe and summarize our previous studies related to the origin and the genetic diversity in Wagyu and Asian cattle.
Japanese Wagyu Breeds
All modern cattle were derived from the wild ancestor of aurochs (Bos primigenius). The aurochs distributed throughout large parts of Eurasia and Northern Africa. The modern cattle are categorized into two subspecies, Bos taurus and Bos indicus. In North Eastern Asia including Mongolia, North China, Korea and Japan, most cattle lack humps and are classified as Bos taurus. The Bos taurus is thought to be domesticated in the Fertile Crescent around 10,000 years ago (Loftus, 1994; Troy, 2001; Chen et al., 2010).
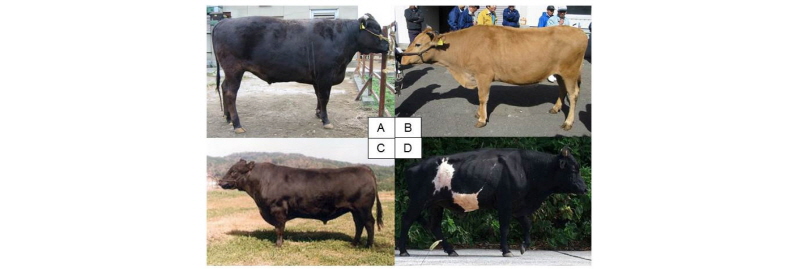
In Japan, we have four breeds of native cattle (Wagyu), Japanese Black cattle, Japanese Brown cattle, Japanese Shorthorn and Japanese Polled. Figure 1 is photos of three breeds of Wagyu and a native cattle of Kuchinoshima feral cattle. All breeds belong to Bos taurus and are beef cattle. Japanese Black cattle is the predominant beef breed (>97%) and is famous as its high-quality meat. Japanese Brown cattle is categorized into two different sub-strains, Tosa-strain in Kochi prefecture and Higo-strain in Kumamoto prefecture. Japanese Shorthorn has been improved by crossbreeding with imported European and American Shorthorn to the indigenous native cattle and now is bred in the northern region of Japan. Japanese Polled has also been improved by crossbreeding with imported Aberdeen Angus and has been maintained in a limited region of Yamaguchi prefecture in Japan. The number of cattle is quite low around 200 heads. These breeds were established by crossing Japanese native cattle with several breeds of European cattle during the mid-19th century to improve the native stock.
In addition to the Wagyu breeds, we have two unique native cattle populations in Japan, Mishima and Kuchinoshima cattle. Mishima cattle has been isolated on Mishima Island for at least 200–300 years and are conserved as a closed colony. Mishima cattle retains the characteristics of native Japanese cattle and were declared a “national natural treasure” in 1928 (Tsuda et al., 2013). Kuchinoshima cattle are feral cattle that originate from the grazing Japanese native cattle from Kuchinoshima Island in the Tokara Island chain of Kagoshima (Figure 1) (Siqintuya et al., 2014). These native cattle are considered to be maintained without any genetic influences from European breeds.
Genetic Diversity of Wagyu Cattle using mtDNA
Mitochondrial DNA (mtDNA) is a powerful tool and D-loop region of the mtDNA has been widely used for studying the origin, genetic diversity, and relationships in cattle (Chen et al., 2010; Loftus et al., 1994; Mannen et al., 1998, 2004; Sasazaki et al., 2006; Troy et al., 2001). The D-loop sequences revealed well diverged major haplogroups T and I in Bos taurus and Bos indicus, respectively. The mtDNA of Bos taurus consists of mainly five sub-haplogroups as T, T1 - T4. Previous studies indicated the genetic diversity and the relationships in Japanese Black cattle (Mannen et al., 1998, 2004), Japanese Brown cattle (Sasazaki et al., 2006), Japanese Polled (Mannen et al., 2017), Kuchinoshima cattle (Mannen et al., 2017) and Mishima cattle (Shi et al., 2002) using mtDNA variations.
At first, Mannen et al. (1998) demonstrated the mtDNA diversity of Japanese Black cattle. In this study, we revealed that Japanese Black cattle has unique mtDNA haplogroup in addition to the other major haplogroups in European breeds. At a later time, the unique mtDNA haplogroup was designated as haplogroup T4 (Mannen et al., 2004). The haplogroup T4 revealed a starburst topology with a center of a haplotype, such as a common, phylogenetically central haplotype with derivative sequences differing by only a few substitutions. Subsequently, we performed the mtDNA analyses for Japanese Brown cattle and Japanese Polled (Sasazaki et al., 2006; Mannen et al., 2017). The mtDNA topology was similar to the Japanese Black cattle.
mtDNA Diversity in Northeastern Native Cattle
We enhanced to the analysis with Korean and Mongolian native cattle, which are representative samples in North Eastern Asia and have analyzed these in conjunction with previously published data from Japanese, Europe, India and Africa (Mannen et al., 2004). Unexpectedly, Mongolian native cattle have Bos indicus mtDNA haplogroup I with adequate frequency (20%). Therefore, we examined gene frequencies of Bos taurus and Bos indicus Y chromosome haplotypes for North Eastern Asian cattle, however, no Bos indicus Y chromosome introgression was observed in Mongolian cattle.
Mongolian cattle have strictly Bos taurus morphological features, as do Japanese and Korean cattle. In addition, Mongolia is located in northern and cooler climate area and this does not offer a selective advantage for the heat-tolerant of Bos indicus cattle. No Bos indicus mtDNA in both Japanese and Korean cattle suggests that the introgression may be a secondary phenomenon, with the earlier cattle in the region being purely Bos taurus. The mtDNA introgression is likely to have occurred in recent millennia.
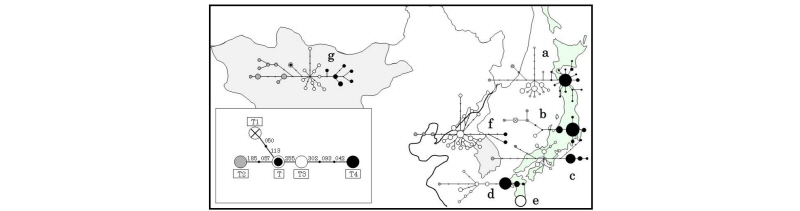
Figure 2.
Reduced median networks of Bos taurus mitochondrial haplotypes in Japan, Korea, and Mongol. The lower left figure panel shows the relationships of the representative five Bos taurus haplogroups T–T4. a: Japanese Black, b: Japanese polled, c: Japanese Brown Kochi strain, d: Japanese Brown Kumamoto strain, e: Kuchinoshima feral cattle, f: Korean native cattle (Hanwoo), g: Mongolian native cattle. This figure was edited from Mannen et al. (2017).
The Bos taurus mtDNA in Korean and Mongolian cattle revealed high genetic diversity. The Korean and Mongolian cattle showed T, T2, T3 and T4 haplogroups, as well as Japanese mtDNA haplogroups. However, these haplogroups did not indicate star burst topology such as Japanese T4 topology (Figure 2). This suggests that continental native cattle have higher mtDNA diversity than that of Japanese cattle.
Origin of Japanese Wagyu by mtDNA Diversity
Generally, ancestor of Japanese cattle is thought to be migrated from North China via the Korean peninsula to Japan around the 2nd century AD and then expanded from west region to all over Japan (Mannen et al., 1998). From our previous studies, all mtDNA of the Japanese Wagyu breeds (Japanese Black cattle, Japanese Brown cattle, Japanese Polled) belonged to common Bos taurus haplogroup T, including T, T1, T2, T3 and T4 (Mannen et al., 1998; 2004; 2017). The central star burst topology of haplogroup T4 in Japanese Wagyu breeds, but was not observed in continental Korean and Mongolian native cattle. A plausible scenario of the Japanese central starburst topology is a result of bottleneck and founder effects in ancient time of cattle migration to Japanese Island. It is likely that only a small number of animals were introduced to the Japanese Islands at that time because of the Japanese sea barrier.
Genetic Diversity of 16 Eurasian Cattle Including Wagyu by SNPs
In our previous study (Yonesaka et al., 2016), we genotyped 117 autosomal SNPs using a DigiTag2 assay to assess the genetic diversity, structure and relationships of 16 Eurasian cattle populations of Bos taurus (Black Angus, Hereford, Japanese Holstein, Hanwoo, Tosa Japanese Brown, Higo Japanese Brown, Japanese Shorthorn, Japanese Polled, Japanese Black and Mongol native cattle) and Bos indicus (native cattle from Laos, Cambodia, Vietnam, Myanmar, Bhutan and Bangladesh). Phylogenetic and STRUCTURE (K=2) analyses showed distinct separation between Bos taurus and Bos indicus populations. Figure 3 illustrated the result of STRUCTURE analysis. This demonstrated the distinct separation between European and Asian populations (K=3). At K=11, each Bos taurus population formed an independent cluster. Mongolian (K=13-16) and Japanese Black (K=14-16) populations showed admixture patterns with different ancestries. Bos indicus populations exhibited a uniform genetic structure at K=2-11, suggesting that there are close genetic relationships among Bos indicus populations. Although we developed and used only 117 SNPs, this study could explain the genetic construction of Asian cattle populations.
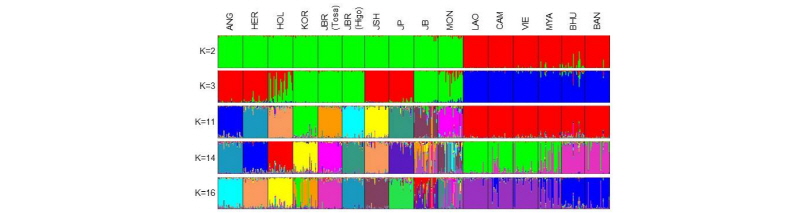
Figure 3.
Genetic structure of 16 cattle populations based on 117 SNPs using STRUCTURE 2.3.4. Each individual is represented as a single vertical line and the proportion of the colored segment represents their estimated ancestry deriving from different populations. ANG, Black Angus; HER, Hereford; HOL, Japanese Holstein; KOR, Hanwoo; JBR (Tosa), Tosa Japanese Brown; JBR (Higo), Higo Japanese Brown; JSH, Japanese Shorthorn; JP, Japanese Polled; JB, Japanese Black; MON, Mongolian native cattle; LAO, Laotian native cattle; CAM, Cambodian native cattle; VIE, Vietnamese native cattle; MYA, Myanmar native cattle; BHU, Bhutanese native cattle; BAN, Bangladeshi native cattle. This figure was edited from Yonesaka et al. (2017).
Genetic Diversity of Kuchinoshima Feral Cattle
Kuchinoshima cattle are unique feral cattle in Kuchinoshima Island in Kagoshima prefecture (Siqintuya et al., 2014). It was reported that a small number of Japanese native cattle were introduced to Kuchinoshima Island in 1918 or 1919 (Hayashida & Nozawa, 1964). Then, some of these cattle escaped to the mountains and became the founder of the Kuchinoshima feral cattle. So far, this population has proliferated naturally in the mountains of Kuchinoshima Island. Kuchinoshima cattle are maintained as small populations (< 200) and have been subject to significant inbreeding.
From our previous studies (Mannen et al., 2017; Saito et al., 2016), the Kuchinoshima cattle have quite low genetic diversity. By the mtDNA analysis, 32 Kuchinoshima cattle have only an identical haplotype belonged to haplogroup T3. In addition, we genotyped 54K SNPs for Kuchinoshima cattle to estimate genetic diversity and genetic structure in Kuchinoshima cattle (Saito et al., 2016). The average minor allele frequency in Kuchinoshima cattle was quite lower (0.089) than those of the other populations of Japanese Black, Japanese Brown and Japanese Holstein (0.181 - 0.251). These results of mtDNA and 54K SNPs chip indicated the extremely low genetic diversity in Kuchinoshima cattle, due to founder effect derived from a few ancestral individuals and genetic drift caused by small population with below 100 animals lasting for a long time in Kuchinoshima Island. In spite, Kuchinoshima cattle still have coat-color variation (Figure 1) and gene variants of functional genes (NCAPG, FASN, SCD, SREBP-1, F11 and MC1R) (Siqintuya et al., 2014).