Introduction
Goats (Capra hircus) are one of the oldest livestock species to have been domesticated, and they spread over a wide range of habitats with a substantial concentration in the tropics and dry zones in developing countries (FAOSTAT, 2006). In Indonesia, they are distributed across the country and usually found in small holdings as mixed flock with sheep. They are kept as an essential resource for meat, milk and fiber. Goat is considered the most prolific of all domesticated ruminants especially in harsh climatic conditions (Yadav and Yadav, 2008).
In goat farming, litter size is a major concern of profitability. Litter size depends on a number factors among which ovulation rate is especially important. So far, goat genetic improvement schemes in Indonesia have involved crossbreeding trials, with examples Senduro, Boerawa, Ettawa Grade and Boerka goats. Also, the use of conventional breeding methods, such as phenotypic information is a simple procedure being expanded. However, since the heritability of litter size is low, it is imperative to explore a suitable candidate gene, which may responsible on prolificacy trait in goats.
Growth differentiation factor 9 (GDF9) belongs to the transforming growth factor β superfamily and plays a critical role in ovarian follicular development and ovulation rate (Elvin et al., 1999; McNatty et al., 2005). The gene is mapped to autosome 5 in sheep (Sadighi et al., 2002). Given the central role of GDF9 in ovulation and reproduction, GDF9 is a good candidate gene for mutations associated with reproductive performance. This gene has been widely studied in humans, sheep, and goats (McNatty et al., 2003). This gene is the last major gene found to be associated with prolific phenotype, in which a polymorphism (FecGH) found in Cambridge and F700-Belclare sheep is responsible for an increased ovulation rate in heterozygotes and sterility in homozygotes, in a way very similar to all BMP15 variants. Later some specific mutations found on this gene have been shown to be associated with different phenotypic effects on litter size in goats (Feng et al., 2011; An et al., 2012; Silva et al., 2010).
The aim of this study was to explore the polymorphism of GDF9 gene as an important fecundity gene in Indonesian goats.
Materials and Methods
Animals and DNA extraction
A total of 77 animals, belonging to nine Indonesian goat breeds (Gembrong = 5, Senduro = 3, Ettawa Grade = 3, Boerawa = 7, Boerka = 25, Kosta = 4, Samosir = 8, Boer = 12, and Kacang = 10) were genotyped in this study. Those animals were reared under the same feeding and management in Indonesia Goat Research Center in Sei Putih, North Sumatera, Indonesia. Approximately 3 ml of blood samples was collected from the jugular vein. The collected samples were transported to the laboratory at 40C before DNA extraction, which was performed using gSYNCTM DNA Extraction Kit (Geneaid, New Taipei City, Taiwan).
Genotyping of GDF9 gene polymorphism
A 462 bp region of GDF9 gene was amplified using a pair of primers, forward (5’-GAAGACTGGTATGGGGAAATG-3’) and reverse (5’-CCAATCTGCTCCTACACACCT-3’), as described by Hanrahan et al. (2004). PCR was performed in SEDI Thermo Cycler in 25 μL reaction containing 2 μL of genomic DNA, 12.5 μL of MyTaqTM HS Red Mix (Bioline, UK), 0.5 μL of each primer (forward and reverse) and 9.5 μL of double-distilled water. The PCR cycling procedures were optimized as follows: initial denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 1 min, 61°C for 1 min and 72°C for 30 s, and a final extension at 72°C for 4 min. The PCR products were visualized on a 1.5% standard agarose gel stained with ethidium bromide and documented using a digital camera.
To genotype all the tested samples, we used SNP g.306G>A (Javanmard et al., 2011; Hanrahan et al., 2004) by PCR-RFLP method using HhaI restriction enzyme (5’...GCG↓C...3’) and SNP g.228C>A using Hpy166II restriction enzyme (5’...GTN↓NAC...3’). A 10 μL of digestion reaction consisted of 5 μL PCR product, 2.4 μL double-distilled water, 2 μL of 10X buffer and 0.6 μL of restriction enzyme. The final volume of mixture was mixed and spinned down for few seconds, then incubated for 5 min at 37°C in dry thermo bath (EYELA, Japan) using both restriction enzymes. Restriction digestion products were checked by electrophoresis using 3% agarose gel in 1X TBE running buffer and staining with ethidium bromide. The 100-bp ladder was used as molecular size marker. In addition, we also used direct sequencing in both directions to confirm the target gene and identify the possible polymorphism within 462 bp of exon 1 in GDF9 gene by 1st BASE DNA Sequencing services.
Statistical analysis
A Chi-square test was performed to test the allelic and genotypic frequencies for Hardy-Weinberg equilibrium. The following mathematical model was:
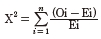
Where, X2 is Chi-square value, Oi is observed frequency, Ei is expected frequency, n is the number of possible outcomes of each event.
Results
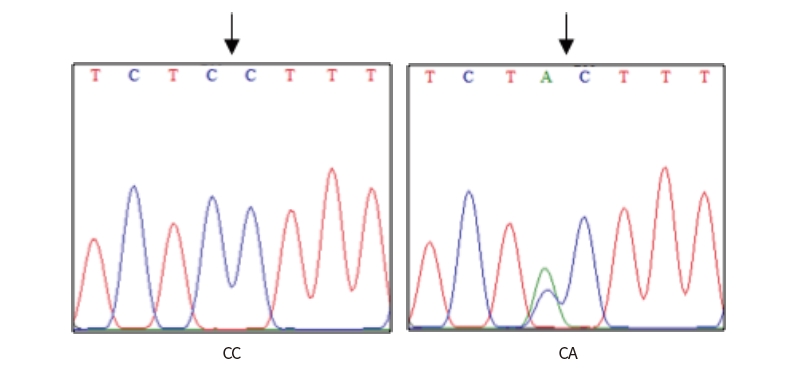
A single fragment of approximately 462 bp nucleotide sequence of exon 1 in GDF9 gene was successfully amplified from each sample of Indonesian goats. PCR products were checked with 1.5% agarose gel electrophoresis and the results were consistent with the target fragments and had good specificity. Two SNPs were identified in this study, SNP g.306G>A and SNP g.228C>A. The SNP g.306G>A was identified based on reference. This missense mutation (CGC to CAC) altered amino acid from Arginine to Histidine, at position 87 of 132 amino acid protein sequences in the exon 1 of GDF9 gene (Figure 1). The SNP g.228C>A was recognized by direct sequencing of 4 samples, representing 4 Indonesia goat breeds (Gembrong, Etawah Grade, Kosta and Samosir), as presented in Figure 2 and 3. The SNP g.228C>A did not induce to amino acid change, resulting leucine amino acid at position 61.
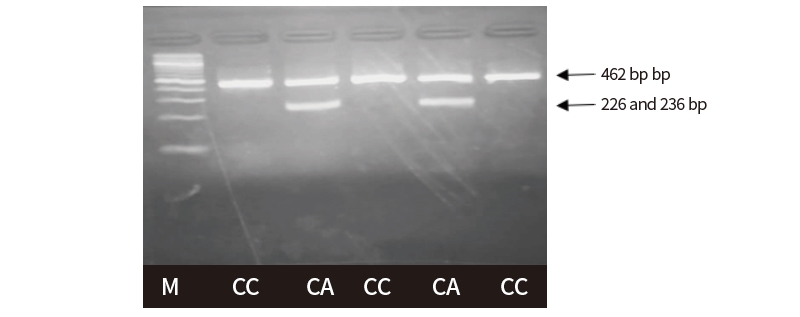
The HhaI restriction enzyme was used in order to detect the polymorphism based on SNP g.306G>A. Agarose gel electrophoresis revealed single type of restriction pattern, consisting of 3 fragments: 52 (not visible), 156 and 254 bp. No polymorphism in exon 1 of GDF9 gene was found among all typed samples and all individuals had GG genotype based on SNP g.306G>A. Digestion of Hpy166II restriction enzyme was carried out in all PCR products to detect polymorphism based on SNP g.228C>A. The results showed two kinds of genotypes, CC genotype (the single band sized 462 bp) and CA genotypes (the three bands, sized 462, 226 and 236 bp), as presented in Figure 4. The success of the Hpy166II (5’...GTN↓NAC...3’) restriction enzyme to digest the PCR products showed that A allele were exist among tested animals. In contrast, the C allele is indicated by the failure of Hpy166II to find a restriction site along the PCR products.
SNP identification of CAPN-1 Gene and Restriction Enzymes
Only one SNP detected (g.4732T>G) on the sequence alignment with three GenBank (AF248054 as a target sequence, NC_032678, NC_037356) (Fig 4). However, four SNPs were found indirect sequencing of Ongole Grade cattle and Kebumen Ongole Grade cattle, namely g.4481A>G, g.4519C>A, g.4554C>T, and g.4631T>C (Fig 5). Two SNPs located in intron 4 (SNPs g.4481A>G and g.4519C>A) and one SNP in intron5 (SNP g.4631T>C). The SNP g.4554C>T was located in exon 4. This SNP have synonymous mutation without amino acid changes.
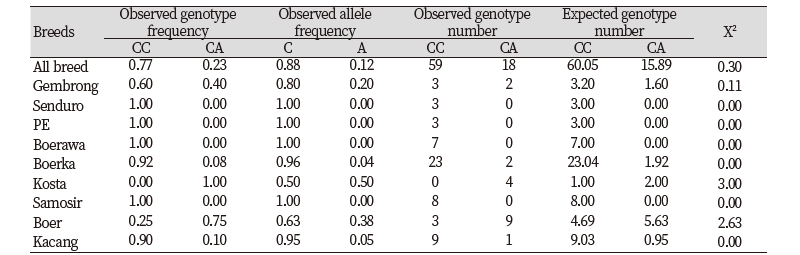
Allele and genotype frequencies of GDF9 gene based on SNP g.228C>A in nine goat breeds are presented in Table 1. The frequency of C allele was higher than that of A allele. The C allele was dominant in all breed tested in this study (more than 50%), except Kosta goat, which has similar frequency for both C and A alleles. The CC genotype had the highest frequency (77%) than CA genotype (23%) in all breed. There was no homozygous AA animals appeared in this study. Only one genotype (CC) was recognized in Senduro, Ettawa Grade, Boerawa, and Samosir goats. On the contrary, all tested Kosta goats was heterozygous animals (CA). Both CC and CA genotypes were detected in samples of Gembrong, Boerka, Boer, and Kacang goats. The CC genotype frequency in Gembrong, Boerka, and Kacang was higher than the CA genotype frequency, which was different in Boer goat. Boer goat had the highest heterozygous CA genotype (75%). All breeds were analysed using Chi-square test to determine the Hardy-Weinberg equilibrium. The result showed that both allele and genotype distribution of all population studied were not deviated from Hardy-Weinberg equilibrium (P>0.05).
Discussion
Prolificacy is one of the most important economic traits in sheep and goats. Heritability of the litter size is low and its improvement by conventional breeding methods is a relatively slow process. Marker-assisted selection, together with traditional selection methods, could gain a better genetic progress in litter size. In sheep and goat, candidate gene markers for litter size have been identified, such as GDF9 (Davis, 2004; Ran et al., 2009; Ma et al., 2010; Dutta et al., 2013; Gazooei et al., 2013).
The GDF9 gene can be considered as a possible candidate gene for increasing litter size in sheeps and goats. Li et al. (2003) identified one single nucleotide mutation (A152G) of GDF9 gene in Hu, Dorset and Suffolk sheep by PCR-single strand conformation polymorphism (SSCP). Hanrahan et al. (2004) reported eight DNA variants in GDF9 of Cambridge and Belclare sheeps including G1 to G8. Feng et al. (2011) identified five nucleotide changes between sheep (according to AF078545) and goats (Jining Grey, Boer, and Liaoning Cashmere) GDF9 gene, which were T114C and G199A at coding base in exon 1, and G423A, A959C and G1189A mutation in exon 2, respectively. Futhermore, the A959C and G1189A nucleotide changes has been reported by Ahlawat et al. (2016) in seven Indian goat breeds. The sequencing results reported by Yan et al. (2010) revealed one mutation (c.792G → A) at exon 2 of GDF9 gene in goats, and this mutation leads to an amino acid change: valine to isoleucine. In the present study, based on SNP g.306G>A, results from reference, we did not find any polymorphism within exon 1 of GDF9 gene by PCR-RFLP among all the investigated animals. This finding was similar in Indian (Ahlawat et al., 2013), Chinese (Ma et al., 2010), Indian Black Bengal (Polley et al., 2009) and Assam Hill goats (Dutta et al., 2013). Indonesian goats are considered as prolific goats and the presence of twinning birth has been recorded in many flocks. An absence of polymorphism, depicted by one type of restriction pattern based on PCR-RFLP, suggested that SNP g.306G>A was not responsible for prolificacy rate in this goat population. In this study, we found a new polymorphism (SNP g.228C>A) of GDF9 in exon 1 region, but this SNP g.228C>A mutation did not induce to amino acid change.
Genotyping using SNP g.228C>A resulted in two genotypes among Indonesian goats, CC and CA. Ahlawat et al. (2015) reported that only two genotypes were recorded based on A959C mutation in exon 2 of Black Bengal goats. Chu et al. (2011) reported two new polymorphisms (c.183A>C and c.336C>T) in caprine GDF9 gene, and showed an association with allele A at G2 locus between GDF9 gene and litter size in Jining Grey Goats. The same result was also reported by Feng et al. (2011), in which does with genotype CC or AC had 0.81 (P<0.01) or 0.63 (P<0.01) kids more than those with genotype AA, respectively. Allele C at 959 locus of GDF9 gene was associated with high litter size in Jining Grey goats. Yan et al. (2010) reported that in Xinong Shaaneng dairy goat and Boer goat, the average litter size in animals with AA genotype was significantly higher than AG and GG genotypes (P<0.01). Owing to the lack of functional data and small population size used in this study, futher studies for association between SNP and prolificacy are necessary for functional validation for this new polymophism (SNP g.228C>A) found in this study.
In this study, the X2 test showed that SNP g.228C>A in the population studied was in Hardy-Weinberg equilibrium (HWE) (p>0.05). This finding was different in Cashmere goat reported by Wang et al. (2018), who reported that SNP g.3905A>C and SNP g.4135G>A loci were not in HWE (p<0.05). An et al. (2012) reported that in Saanen and Guazhong breeds, three SNP loci were in Hardy-Weinberg disequilibrium,indicating that the population studied might be affected by selection, mutation or migration.