Introduction
Ongole Grade cattle are the result of a cross between local cattle from Java and Ongole cattle from India that have an important role in meeting beef demand in Indonesia (Astuti, 2004). The qualitative characteristics of Ongole Grade cattle based on Indonesian National Standards (SNI 7356: 2008) are white-gray fur, fur around black eyes, large body, large hump, neck and short horns (Anonymous, 2008). Another Ongole grade namely Kebumen Ongole Grade cattle, is a result of crossing between Java, Ongole and Brahman cattle (Nugraha, 2014). Kebumen Ongole Grade cattle have special characteristics with a black muzzle, black tail fan, black color around the eyes, long and hanging ears and has a large hump and dewlap (Ngadiyono et al., 2017). The population of beef cattle in DIY (Yogyakarta Special Regency) in 2016 is ranked 12th at the national level with Ongole Grade cattle composing 40% of the total population of beef cattle (Ditjennak, 2016). The population of beef cattle in Kebumen Regency was higher, with it composing 90% of the total population (Department of Agriculture and Animal Husbandry of Kebumen Regency, 2011). In order to fulfill the demands and tastes of consumers for quality beef, several efforts such as analysis of meat quality and identification of superior livestock should be conducted. One of the methods to improve meat quality is through molecular approach with DNA markers of the gene encoding the meat quality as a selection tool.
FASN is a gene that encodes the quality of meat, including carcass weight and fat content. The FASN gene is responsible for the activity of FASN enzymes and fatty acid synthesis. Zhang et al. (2008) identified SNP in exon 39 (g.17924G>A) which resulted in changes amino acids from threonine to alanine, and was associated with fatty acid composition. In addition, variations in 5 locations (g.15531C>A, g.16907T>C, g.15603G, g.17250–17251A>T in del>, and g.17924G>A) have a significant relationship with the percentage of C14: 0 on the cow adipose tissue and milk fat (Morris et al., 2007). Fat metabolism and the characteristics of obesity have been associated with FASN expression or polymorphism in cattle (Abe et al., 2008). Studies in humans show an increase in FASN expression in adipose tissue from obesity subjects and there is an SNP association in FASN with its expression in cases of obesity (Berndt et al., 2007; Schleinitz et al., 2010). Rempel et al. (2012) found that there were SNP in the genotypes of FASN markers that were associated with growth traits includes final body weight and carcass weight. Maharani et al. (2012) obtaining SNP on the genotypes of FASN markers on fatty acid composition in Hanwoo cattle which revealed this marker had a significant effect on the fatty acid composition.
CAPN-1 gene, encoding protease cysteine which is the primary enzyme in the postmortem tenderization process, is responsible for the breakdown of the myofibrillar protein associated with meat tenderness (Chung et al., 2014). The coding gene for 1-calpain (CAPN1) is located on cattle chromosome number 29, considered a strong functional candidate for beef tenderness (Curi et al., 2009). Some SNPs have been developed for the CAPN-1 gene and these markers have been reported to be associated with meat tenderness in Bos Taurus (White et al., 2005). Two non-identical Single Nucleotide Polymorphisms (SNPs) in the CAPN1 gene have been associated with the traits of meat quality in Bos Taurus (Page et al., 2004). CAPN316, located on exon 9, and CAPN530, is located on exon 14, which is responsible for substitution of amino acids p.Gly316Ala and p.Val530Ile. White et al. (2005) have identified markers in the CAPN1 gene on Bos Indicus. The results showed a separation of CAPN4751 polymorphism (AF_248054.2: g.6545C>T), which was located at intron 17, and there was a significant relationship with meat tenderness in the Bos Indicus, Bos Taurus and the cross of Bos Indicus and Bos Taurus.
Identification SNP of FASN and CAPN-1 genes in this study was conducted on Ongole Grade and Kebumen Ongole Grade cattle. SNP detected will be suggested to be used for genotyping and genetic diversity analysis of Ongole Grade and Kebumen Ongole Grade based on FASN and CAPN-1 gene.
Materials and Methods
Meat Samples and DNA Isolation
The meat samples were collected from twenty-two Ongole Grade cattle in Giwangan slaughter house Yogyakarta and sixteen Kebumen Ongole Grade cattle were collected from slaughter house in Kebumen. The meat samples were taken from longisimuss dorsi which was a part of sirloin. The longisimuss dorsi samples were collected for Genomic DNA isolation using SYNCTM DNA Extraction Kit (Geneaid, Taiwan).
Target Gene Amplifications
Primer used for PCR amplification of FASN is forward: 5 '-TCTTCACAGAGCTGACGGAC-3' and reverse: 5 '-GGAGGAAGAGCTGTTGCAGT -3' with GenBank Acc.No.: NC_032668.1 (Maharani et al., 2012) and CAPN-1: forward: 5'- AGTGAGTAGAAAGCCCTCCC -3 'and reverse: 5'- AGGTAAACAGCTCAGCACAGAC -3' with GenBank Acc.No. : AF248054 (Chung et al., 2014). Amplifications for FASN were performed under the following conditions: 10 minutes for 94oC, 35 cycles of 30 seconds at 94oC, annealing 61oC for 30 seconds, extension 72oC for 30 seconds and the final extension 72oC for 10 minutes. Amplifications for CAPN-1 run with conditions: pre-denaturation occurred at 95oC for 2 minutes, denaturation 94oC for 1 minute, annealing 65oC for 1 minute, extension 72oC for 90 seconds and the final extension 72oC for 10 minutes in 35 cycles. The products were visualized in 1.5% agarose gels and stained with ethidium bromide then visually with Ultra Violet (UV) light and documented using a digital camera.
Determination of SNP and restriction enzymes
SNP determination was carried out using the sequence alignment method of various types of cattle based on the FASN and CAPN-1 GenBank from www.ncbi.nlm.org and alignment sequences of Ongole Grade and Kebumen Ongole Grade samples. Sequencing was performed by 1st Base PT Genetika Science Indonesia. Determination of restriction enzymes was performed using Bioedit and Nebcutter software (http://nc2.neb.com/NEBcutter2/).
Results
Identification SNP of FASN gene and Restriction Enzyme
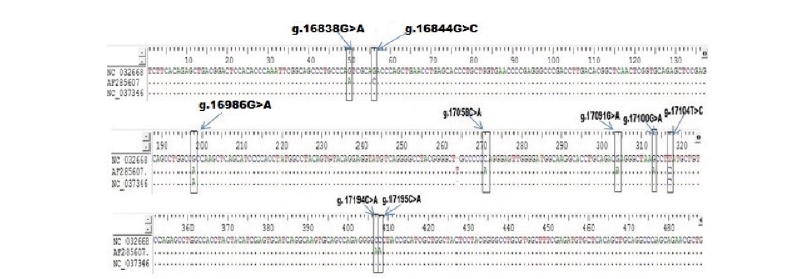
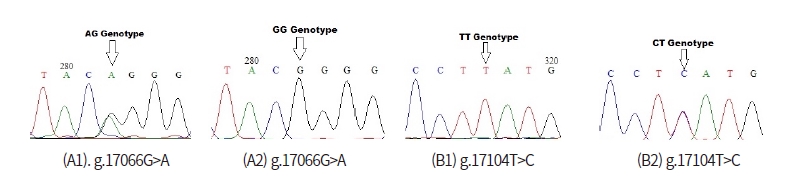
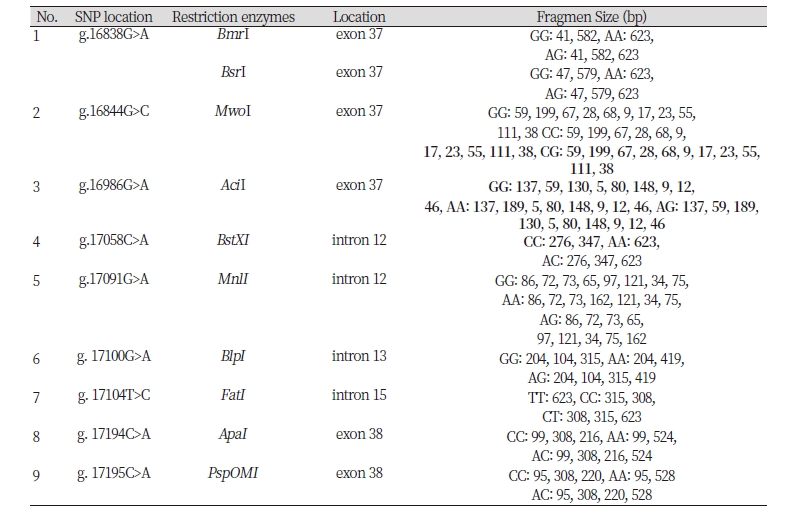
In order to detect SNPs of FASN gene, alignment based on three GenBank sequence (NC_032668 as target sequence, AF285607, NC_037346) from Bos Taurus cattle and direct DNA sequencing of Ongole Grade and Kebumen Ongole Grade cattle were performed. As a results, nine SNPs in the FASN gene based on the GenBank’s alignment sequences have been identified namely SNP g.16838G>A, g.16844G>C, g.16986G>A, g.17058C>A, g.17091G>A, g.17100G>A, g.17104T>C, g.17194C>A, and g.17195C>A (Fig 1). Moreover, all of the nine SNPs have a non-synonymous mutation with the amino acid changes shown in Fig 2. Based on the direct sequencing, two SNP were found in intron 27, namely SNP g.17066G>A and SNP g.17104T>C (Fig 3). SNP g.17104T>C found in both GenBank alignments and direct sequencing. The SNPs g.17066G>A and g.17104T>C were not induce to amino acid changes.
Restriction enzymes are determined based on the SNP identified in GenBank alignment and SNPs found in this study. The list of restriction enzymes and their fragment size based on the results of GenBank alignment shown in Table 1. One recommended enzymes for genotyping the Ongole Grade cattle and Kebumen Ongole Grade cattle in SNP position g. 17104T>C was FatI enzyme. However, no restriction enzyme detected in SNP g.17066G>A.
SNP identification of CAPN-1 Gene and Restriction Enzymes
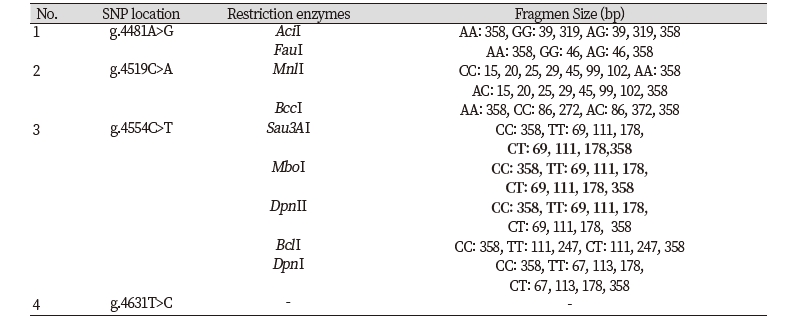
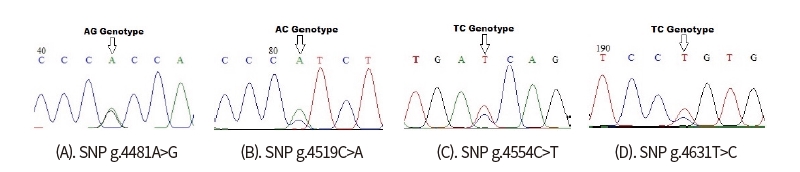
Only one SNP detected (g.4732T>G) on the sequence alignment with three GenBank (AF248054 as a target sequence, NC_032678, NC_037356) (Fig 4). However, four SNPs were found indirect sequencing of Ongole Grade cattle and Kebumen Ongole Grade cattle, namely g.4481A>G, g.4519C>A, g.4554C>T, and g.4631T>C (Fig 5). Two SNPs located in intron 4 (SNPs g.4481A>G and g.4519C>A) and one SNP in intron5 (SNP g.4631T>C). The SNP g.4554C>T was located in exon 4. This SNP have synonymous mutation without amino acid changes.
Determination of restriction enzymes was carried out in accordance with SNP results from GenBank alignment and SNP identified from direct sequencing of both sample breeds. Restriction enzymes based on the results of GenBank alignment with SNP g.4732T>G, namely DpnII, Sau3AI, MboI, DpnI, TaqI, PvuI, ClaI, and BspDI. The restriction enzymes and the fragment size of SNP detected in Ongole Grade cattle and Kebumen Ongole Grade cattle shown in Table 2.
Discussion
The FASN enzyme consists of two multifunctional polypeptides identical to the three catalytic domains in N-terminals (b-ketoacyl synthase, malonyl acetyl transferase, and dehydrase). The catalytic domain is separated by 600 residues from the C-terminal region formed by four other domains (enoyl reductase, b-ketoacyl reductase, acyl carrier protein, and thioesterase). The FASN enzyme is one of the most complex multifunctional enzymes. This enzyme has the catalytic component needed to catalyze a series of 37 reactions that lead to the formation of palmitic acids from acetyl-CoA and malonyl-CoA (Asturias et al., 2005). The mechanism of action of the FASN gene is by catalyzing the synthesis of palmitate from acetyl-CoA and malonyl-CoA. One by-product of the pyruvate reaction is acetyl-coenzyme (CoA); together with malonyl-CoA, it becomes a substrate for FASN, which catalyzes the biosynthesis of palmitic fatty acids in nicotinamide adenine dinucleotide phosphate - reduced (NADPH) - depending on the reaction. Palmitate is then conjugated with other proteins or converted to other fatty acids and lipid complexes which are essential for (i) synthesis of lipids and membrane structures, such as lipid rafts, (ii) protein modification and localization function, and (iii) receptor localization and signaling main pathways oncogenic conditions such as PI3K / AKT / mTOR pathway (Jones and Infante, 2015).
In this study, there were 2 polymorphisms detected in the 623 bp fragment of FASN gene of Ongole Grade cattle and Kebumen Ongole Grade cattle. This study discovered 8 different SNPs and one similar SNP FASN gene in 5’UTR region when we compared with the direct sequence and 3 GenBanks from NCBI. This study revealed that sequence of FASN Ongole Grade cattle and Kebumen Ongole Grade cattle was a similiar with another sequence from Bos Taurus cattle (Maharani et al., 2012). The FASN gene in this region may be used to distinguish Ongole Grade cattle and Kebumen Ongole Grade with other species. Both of two SNPs based on direct sequencing (g.17066G>A and g.17104T>C), were located in non-coding region. Non-coding sequences are important for regulatory functions. An untranslated region that controls their translation, degradation, and localization include stem-loop structures, upstream initiation codons and open reading frames, internal ribosome entry sites and various cis-acting elements that are bound by RNA-binding proteins (Mignone et al., 2002). Five SNPs from GenBank alignments (g.16838G>A, g.16844G>C, g.16986G>A, g.17194C>A, and g.17195C>A) have amino acid mutation: valine to isoleucine, aspartic acid to histidine, arginine to histidine, proline to threonine, proline to histidine, respectively. Therefore the SNPs may control the genotype and phenotypic expression.
An attempt at determining restriction enzymes with SNP in Ongole Grade cattle and Kebumen Ongole Grade cattle obtained results that only SNP g.17104T>C can be digested by restriction enzymes, namely FatI enzyme. Therefore, SNP g.17104T>C can be used in the stage of determining the genotype of livestock by PCR RFLP method. The enzyme can be digested the gene target and revealed TT genotype with similar size of the gene target (623 bp). The homozygote CC having fragment sizes: 308 bp and 315 bp, and TC genotypes revealed having fragment size: 308 bp, 315 bp, and 623 bp. Based on previous studies, there are several SNPs detected in FASN gene that are associated with meat quality. Maharani et al. (2012) found SNP g.17924G>A with a MscI restriction enzyme associated with myristic acid, which is one of the fatty acid compositions of meat.
Calpain is a group of cysteine protease enzymes that require calcium ions for their activities. CAPN-1 has a broad sub-network and reflects a large number of targets, such as vimentin (VIM), collagen bonds (COL1A1), or desmin (DES). Calpain has implications for various calcium-dependent cellular processes such as apoptosis (Perrin and Huttenlocher, 2002). Functional analysis of CAPN-1 tissue involves developmental process (23%), regulation and apoptosis (19%), an organization of cellular components and biogenesis (15%), and response to external stimuli (10%). Mechanisms such as development and stress, apoptosis, glucose metabolism, and muscle contraction contribute to the proteolytic pathway of CAPN-1. This allows the interaction in the muscle to explain the relationship of protein to tenderness. Stress plays an important role in forming SUMO4, H2AFX, and HSPs and involves certain cells so that cells can survive with stressful conditions by maintaining metabolic functions and structural proteins (Guillemin et al., 2011).
This study in CAPN-1 gene discovered one SNP based on the alignment of three sequences from GenBank in NCBI. However, it was found that 4 polymorphisms detected in the 358 bp fragment of CAPN-1 gene in Ongole Grade cattle and Kebumen Ongole Grade cattle. This reveals that sequence of CAPN-1 from direct sequencing was a difference compared with another sequence from Bos Taurus cattle. The SNPs in CAPN gene in this region may be used to recognize Ongole Grade cattle and Kebumen Ongole Grade with other species. Three SNPs in this study were located in noncoding region and 1 SNP were identified in coding region in exon 4 and were not induced by amino acid changes.
Restriction enzymes based on SNP detected in Ongole Grade cattle and Kebumen Ongole Grade cattle exhibited that 3 SNPs can be digested by restriction enzymes, and 1 SNPs (SNP g.4631T>C) can not be digested by enzyme restriction. In previous studies, there are several SNPs from the CAPN-1 gene that are related to meat quality. Chung et al. (2014) discovered SNP g. 4558A>G with MboI restriction enzyme, which is responsible for the breakdown of a myofibrillar protein associated with meat tenderness.
In conclusion, we detected two synonymous polymorphisms of FASN gene in Ongole Grade cattle and Kebumen Ongole Grade cattle in the location g.17066G>A and g.17104T>C. Restriction enzymes FatI may be used for genotyping Ongole Grade cattle and Kebumen Ongole Grade cattle by applying PCR-RFLP method in future researches using SNP g.17104T>C. The study of CAPN-1 gene in both cattle detected three synonymous polymorphism in location g.4481A>G, g.4519C>A, and g.4631T>C and one non-synonymous polymorphism in location g.4554C>T in exon 4. Restriction enzymes AciI, FauI, MnlI, BccI, Sau3AI, MboI, DpnII, DpnI, and BclI may be used for genotyping Ongole Grade cattle and Kebumen Ongole Grade cattle using PCR-RFLP method in the future research.