Introduction
Mongolia has a long history of breeding domestic animals across its vast territory. Mongolia system of pastoral herding consists five kinds of domestic animals and dates back more than 3,000 years. Unfortunately, we could not find genetic research into the origins of Mongolian domestic animals. However, there were some short archeological reports on the history of domestic animals (Navaan, 1975; Sanjmyatav, 1982; Ser-odjav, 1987; Odsuren et al., 2015). These papers were based on archeological evidence, such as petroglyphs, human burial, and other archeological findings. The authors demonstrate that in central Asia (including Mongolia) ancient inhabitants began raising domestic animals in the middle Neolithic age or New Stone age (10,000 - 7,000 BC) (Navaan, 1975; Ser-odjav, 1987).
Mongolian domestic animals are well adapted to harsh weather conditions; however, their productivity is not good than commercial livestock. They provide a significant source of food and raw materials (e.g., cashmere) for the national economy which are contributing 19% to the total Gross Domestic Product (GDP). According to government statistics, in 2017 an estimated 61.5 million head of goats, sheep, cattle, horses and camels lived in Mongolia (NSOM, 2017). Foreign breeds of sheep, goat and cattle were introduced to Mongolia during the Soviet period (between 1950s-1990s) from Socialist countries as part of an animal breeding program controlled by Russian veterinarians in a collective management system. Today, livestock owners control the breeding, but the collective systmen provided opportunities to improve animal performance of Mongolian livestock. Superior animals were selected based on their phenotypic performances to improve their productivities.
The People’s revolution has won in 1924 (from China), after revolution by end of 1920s, the first attempt to livestock herding collectives was occurred (under Russian pressure). Unfortunately, because of poor arrangement and plan, the most of herders these herders left collectivization before 1950s (after 1950s, the government paid much attention to rebuild well-arranged collectives with strongly encouraged people). During collectivization, there were advantages and disadvantages, in example, herders were paid heavily tax, in contrast, at this time collectives permitted some ownership of livestock to private herders. The collectivization was aimed to build up a excess of livestock products to provide urban populations by food, both inside and outside of Mongolia (Reading et al., 2006). By after 1990 democratic revolution, collectives were collapsed with the breakup of the command economy. The collectives distributed their equities to in two aspects in 1991 and 1992, large herds of livestock were also distributed among the collective former members (Bruun, 1996). Recently, few formal, large-scale breeding programs exist for Mongolian domestic animals. Instead, small, herders manage about 61.5 million domestic animals across more than 80% of Mongolia (>156 million ha).
Previous population genetics studies focused primarily on domestic Bactrian camels (Camelus bactrianus), a species that is rapidly declining across its range. Additional studies have examined the Mongolian native horse. The majority of Bactrian camel populations inhabit Mongolian and Chinese deserts; therefore, genetic studies focused on comparing the characteristics of Mongolian and Chinese camel population genetics (Jianlin et al., 2004; Chuluunbat et al., 2014; Ming et al., 2015, 2017; Yi et al., 2017). With respect to the Mongolian native horse (Equus f. caballus), its population is one of the largest and most valuable genetic resources for the global horse population, and likely resulted from the vast expansions of Mongolian Empires under Attila (5th century) and Genghis Khan (13th century) (Bjornstad et al., 2002). A few population genetics studies conducted for the Mongolian horses which examined genetic relationships between Korean, Japanese and Norwegian horses (Kim et al., 1999; Bjornstad et al., 2002; Choi, 2006; Kakoi et al., 2006). In contrast, only one to two studies have been conducted on domestic sheep, goats, and cattle.
Origin of Mongolian domestic animals
Data on domestic animal domestication first came from osteometry and morphometry evidence collected in archeological sites (Vigne et al., 2005). More recently, extensive genetic studies complimented these data (Zeder et al., 2006) using both modern and ancient samples, thus producing more precise scenarios of the domestication process. The first archaeological evidence of the domestication of sheep, goats and cattle species goes back to around 10,500 BCE in the Fertile Crescent. It seems that goats and sheep were domesticated first (Peters et al., 2000; Zeder et al., 2000), immediately followed by cattle (Helmer et al., 2005).
We review some of the archeological evidence important for understanding Mongolian (indeed Central Asian) domestic animals. Since ancient times, Mongolians have been rearing five domestic animals that playing significant roles in providing food, clothing, and transportation. Thus, Mongolians traditionally believe that their domestic animals originated in their land, and that the domestic horse originated from the Przewalski’s horse Equus ferus przewalskii, the domestic Bactrian camel from wild Bactrian camel (Bactrianus ferus), domestic sheep from argali (Ovis ammon), and the domestic goat from Siberian ibex (Capra sibirica) (Navaan, 1975). However, there was no genetic researches for exploring the origin of domestic animals in Mongolia. Instead, some archeological publications mentioned the history of Mongolian domestic animals (Navaan, 1975; Sanjmyatav, 1982; Tseveendorj, 1985; Ser-odjav, 1987; Odsuren et al., 2015). These previous reports were based on archeological evidence, such as petroglyphs, human burial, and other archeological findings. Tseveendorj et al. (1985) mentioned that animal domestication resulted from a long-term, successful hunting system. And some scientists suggest that animal domestication in Asia and Mongolia began in the Neolithic Era or New Stone Age. Moreover, historical monuments (ceremonial burial of horned skulls of cattle, numerous horse bones) and petroglyphs (patterns of horsemen and cattle) that belong to Neolithic Age indicate that horses and cattle were independently domesticated by central Asian human communities (Odsuren et al., 2015). The important petroglyph “Moojoo” was discovered in mid-1990s. This petroglyph improved our understanding of the history of animal domestication, with more than 100 rock art depictions (18 ibex, 15 cattle, 2 wolves, 2 hunters and more). Based on the painting method used to produce this petroglyph, Tseveendorj (1985) concluded that the origin of Mongolian domestic animals occurred in the VII century BCE (approximately 9,000 years ago).
The Bronze Age is recognized as a period of rapid development in human economy and agriculture throughout the world. Sanjmyatav (1982) discovered the “Chuluut” petroglyph of the Bronze Age that contains horse, cattle, and camel images. He reported that during Bronze Age (including the end of the Neolithic) approximately 3,300-1,200 BCE, ancient people already kept and reared livestock using classic animal husbandry in the territory of Mongolia. In addition, numerous human belongings, such as clothes made of skin, ropes made of wool, and ornaments made of horn were discovered in human burials. Finally, images of hunting strategies, herding livestock, and transportation by horse & cattle are well represented in several petroglyphs from the Bronze Age (Sanjmyatav, 1982).
Population genetics studies on Mongolia domestic animals
Genetic studies on domestic Bactrian camels, Camelus bactrianus
The majority of the population of domestic Bactrian camels inhabits the desert and semi-desert areas of southern and western Mongolian and northern and western China, while small populations remain in Russia and Kazakhstan. For understanding of domestic camel’s domestication is still remains incomplete, while domestication pattern of the other livestock species is well understood (Chuluunbat et al., 2014). Recently conducted studies suggested that camel domestication was taking area covering southern Mongolia and northern China approximately 6,000 YB (Peters and Von den Driesch 1997; Trinks et al., 2012; Burger, 2012). Previous studies suggested that based on modern archeological and genetic evidence the wild Bactrian Camel and its domestic type have a same common ancestor, which may have extinct long time ago before humans began to pay attention to this useful animal (Ji et al., 2009). In addition, the relationship between domestic and wild camels (Camelus ferus), genetic studies have revealed high divergence at their mitochondrial sequence level (Ji et al., 2009; Silbermayr et al., 2010a) and nuclear loci (Jirimutu et al., 2012), excluding the wild Bactrian Camel as an ancestor of modern domestic populations. Most previous studies (Peters and Von den Driesch 1997; Trinks et al., 2012; Burger 2012; Chuluunbat et al., 2014) focused on mitochondrial DNA variation, while only one study (Jianlin et al., 2004) used microsatellite markers for camel population genetics research that including small Mongolian samples. Genetic diversity, calculated as genetic diversity parameters; mean number of alleles (ranged from 3.58 to 4.75) and expected heterozygosity (He= ranged from 0.526 to 0.602), were similar in all populations. Genetic distances (DA) indicate closer genetic relationships between populations within each country (mean DA=0.064 for Mongolian, and 0.052 for Chinese populations) than between the Chinese and Mongolian populations (DA= 0.129) (Jianlin et al., 2008).
Chuluunbat et al. (2014) reported that during las two decades, Mongolian Bactrian camel’ s population size has dropped from about 700,000 to less than 400,000 head of animals. The effects of such a severe decline might impact future sustainable utilization and conservation of this important livestock species. Chuluunbat et al. (2014) also evaluated genetic diversity among Mongolian camel populations. Within a 83-mt-DNA sequence, authors detected 10 polymorphic sites, representing six previously described haplotypes (H1-H5; Silbermayr et al., 2010a) and nine novel haplotypes. Among four studies populations (Galbiin-Gobin Ulaan, Haniin Hetsiin Huren, Hos Zogdort and Mongolian native camel), haplotype diversity (Hd) and nucleotide diversity (Pi) ranged between 0.600 and 0.020, and from 0.0011 to 0.0032, respectively (Table 1). In comparison, Ming et al., (2015) performed variations in their Cytochrome b gene sequence variation in all Chinese domestic Bactrian camel breeds, and camel breeds from Mongolia and Russia, finding that diversities within Chinese population (Pi: 0.00084, Hd: 0.679) was slightly higher than its Russian (0.00068, 0.667) and Mongolia (0.00060, 0.582, respectively) populations had (Ming et al., 2015). The very low level of genetic differences observed among Mongolian domestic camel populations (Chuluunbat et al., 2014), while no geographic structuring not to existed among Cytb sequence haplotypes in domestic Bactrian camel breeds from Mongolian and Chinese (Ming et al., 2015). Moreover, strong gene flow endured between domestic camel breeds during their history. It is likely results of silk-road, pilgrims, armies, merchants and nomads were used numerous number of domestic camels to transport their properties and army supplies between geographically distinct areas across in territories of China and Mongolia (Ming et al., 2015; 2017).
Recently, 16 maternal (cytochrome b [Cytb] and oxidoreductase chain 1 [ND1] genes) haplotypes (including wild Bactrian camel H14) were revealed among all Chinese, Russia and Mongolian domestic camels, and phylogenetic analyses showed that clearly distinct two major haplogroups into wild and domestic (Chuluunbat et al., 2014; Ming et al., 2015, 2017; Yi et al., 2017). The main mitochondrial haplotypes (H-1, H-3, and H-4) were shared by domestic Bactrian camel’ populations in China, Mongolia, and Russia, the data demonstrate the geographical structuring and apparent genetic admixture among the camel breeds or strain from these three countries (Yi et al., 2017).
Genetic studies of the domestic horse, Equus ferus
During the late Pleistocene ~ 11,000 year before (YB), the herds of wild horses were common in the open steppe of North America and Eurasian (Clutton-Brock, 1999). Some studies highlighted that there is a substantial level of genetic variability in Mongolian horse populations than other region (Nozawa et al., 1998; Bjørnstad, 2001). Further, based on previous genetic diversity findings, some studies suggested that Mongolian steppe is can consider as one of the main areas where domestication took place for horse, and these areas display high levels of genetic unevenness and have a greater probability of acting as gene pool than other countries’ population (Zohary, 1996; Vila et al., 2001).
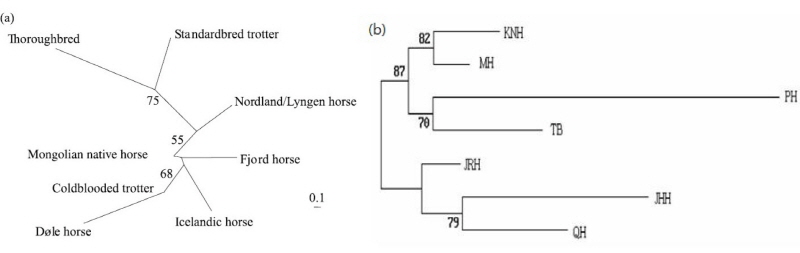
Previously, population genetics research into domestic horses (Kim et al., 1999; Bjornstad et al., 2002; Choi, 2006; and Kakoi et al., 2006) tend to compare their datasets with Mongolian domestic horse samples. The high diversity (Ho = 0.794; He= 0.820) of Mongolian horses likely resulted from vast Mongolian Empires under Attila (II-I century before common era [BCE]) and Genghis Khan (1206- ~1360) that distributed Mongolian horses in wide area across Eurasia during their conquest (Bjornstad et al., 2002). Bjornstad et al. (2002) has investigated the genetic distances between northern Mongolian horses (62 samples) and European breeds using microsatellite genotyping. All the northern European breeds had considerably close genetic distances to the Mongolian horse than distances to the thoroughbreds and Standardbreds. In the phylogenetic tree, the Mongolian horse clustered with the northern European breeds (Figure 1A) (Bjornstad et al., 2002). The genetic distances between Mongolian native horses and Norwegian breeds (DA=0.214) were comparable to the distances found between the Norwegian breeds and the Icelandic horse (DA=0.207), perhaps diverged about 1,000 YB. The Icelandic horse also displayed short genetic distances to Mongolian native horses (DA=0.150) (Bjornstad et al., 2002).
Furthermore, Choi et al., (2006) investigated Korean native horses’ (KNH) genetic diversity and compared it with other horse populations, including Mongolian native (MH), Japan Hokkaido (JHH), Jeju racing (JRH), Quarter horse (QH) and Thoroughbred horse (TB). They used 19 samples from MH that they considered the ancestral breed for KNHs. The expected heterozygosity within breed ranged from 0.636 ± 0.064 (JHH) to 0.809 ± 0.019 (MH) (Choi et al., 2006). Similarly, Kakoi et al., (2007) and McCue et al., (2012) also found that Mongolian horse (n = 21) populations had the highest genetic diversity values (Observed Heterozygosity [Ho] = 0.794, Expected Heterozygosity [He] = 0.820) among seven Japanese and three European breeds. The smallest DA (0.1255) and Ds (0.1566) genetic distances were observed between KNHs and MHs and the largest DA (0.8234) and DS (1.9215) distances were observed between Przewalskii’s horse and the Japanese Hokkaido horse (Choi, 2006) (Figure 1B). Choi (2006) also reported more polymorphic loci in MH than other horse breeds had, this finding demonstrate the MH have retained the big amount of genetic variation of all populations studied. In addition, Mongolian horses owned all the alleles that found in the Korean native horse, with omission at three MS marker (ASB2, HMS3 and HMS7) out of 11. The relatively close genetic relationships demonstrate the KNH is descended from the ancestral populations of MH. Furthermore, Tozaki et al. (2003) performed phylogenetic study for Thoroughbreds, some Japanese horse, and four Asian horse populations (including Mongolian) using microsatellite marker (MS). This study highlighted that Korean peninsula was one important transits corridor for Japanese native horses migration that descended and migrated from Mongolian horses.
Genetic studies of domestic goats, Capra aegagrus hircus
Domestic goats (Capra hircus) have played a significant role in the early time of their domestication, especially for Neolithic agricultural revolution (provides useful products: meat, fiber and milk). The origin of domestic goats has been investigated in several studies particularly using mitochondrial markers (Takada et al., 1997; Luikart et al., 2001; Sultana et al., 2003; Meadows et al., 2007), these studies suggested that this useful domestic animals were probably first domesticated in the Fertile Crescent region of the Near East approximately 10,000 YB.
In Mongolia, goats are important domestic animals, they produce one of the major export products, cashmere beside of their milk and meat (Mongolia is the world’s second largest cashmere producer behind China). Mongolia is producing about 30% of the world’s cashmere demand. Several goat strains are recognized in Mongolia with common names that mostly reared border area (Takahashi et al., 2008). We could find only one genetic diversity study for some Mongolian goat strains (Takahashi et al., 2008), which have lack of native goat sample. However, several genetic studies have been conducted on goat populations in Inner Mongolian, China (Liu et al., 2006; Bai et al., 2006; Di et al., 2010; Wei et al., 2014). Takahashi et al. (2008) investigated the genetic diversity and relationships among some Mongolian goat strains using 10 microsatellite markers, but there was no results for Mongolian indigenous native goat populations. The number of alleles per locus in the eight populations (Bayin-Delgar, Ulgii-Red, Bural, Sumbar, Zala-jinst tsagan, Erchim, Dorgon, and Gobi Gurwan-Saikhan) of goat varied between 7.9 and 9.5, whereas the effective number of alleles ranged from 3.8 to 4.6. The average He and Ho values 0.669 to 0.730 and 0.719 to 0.746, respectively (Takahashi et al., 2008). Genetic differentiation, or Watterson’s theta based on number on segregating sites (θs), for Mongolian goat populations ranged from 0.004 to 0.027, with a mean value of 0.017, suggesting that a high level of gene flow endures between these populations (Takahashi et al., 2008). Surprisingly, the high level of genetic diversity observed within Mongolian goat populations (Ho= 0.669-0.730; He= 0.719-0.746), while exceptionally close genetic relationships also observes among these populations (DA= 0.062). The data suggest that Mongolian goat populations still stay at status of semi-wild genetic structures and have not reached the status of breeds yet. These data might be result of a long history of nomadism across the country and the relatively short history of goat breeding in Mongolia (recently there are no breeding program). Unfortunately, these data did not represent the total population characteristics of goats in Mongolia, with lacking native goat samples.
For other population genetic studies that conducted worldwide, they had identified at least four major mt-DNA lineages for domestic goat (Luikart et al., 2001; Sultana et al., 2003; Chen et al., 2004) with very weak geographic among goat populations, at Control Region (CR) of mt-DNA. For example, mt-DNA types from the main lineage-A was found in all countries and breeds, while lineage-B was found across much of Asia, including India, Pakistan, Mongolia and Malaysia. Representatives of lineage-C was found as far away as Switzerland, Mongolia and Slovenia, when lineage-D was rare and only observed among local goats in Indian and Pakistani. Interestingly, Luikart et al., (2001) found closely related mt-DNA types (different at 1-4 substitutions) in distant locations; Mongolia, Denmark, Ukraine and Portugal. According previous reports, we can easily summarize that Mongolia is one of the few countries that host A, B and C lineages (excluded lineage D). Furthermore, Luikart et al., (2001) also found closely related animals from Mongolia and Ukraine, likely the result of a high level of crossbreeding between introduced foreign breeds from Soviet Union Countries (including Ukraine) with Mongolian indigenous native goats during the Soviet period.
Genetic studies of domestic sheep, Ovis aries
Archaeological and molecular genetic evidence suggests that the wild direct ancestor for domestic sheep (Ovis aries) was probably Ovis orientalis, and domestication occurred approximately 11,000 YB in the Fertile Crescent region (Zeder et al., 2008). The sheep domestication’s genetic history had been previously investigated using maternally inherited mt-DNA in modern sheep breeds (Niemi et al., 2013). Whereas, Mongolians traditionally believed that Mongolian domestic sheep originated from Argali sheep (Ovis ammon); however, Luo et al. (2005) were not found evidence of the contribution of Argali sheep to the maternal origin of both Mongolian and Chinese domestic sheep.
We located only one population genetics study (Luo et al., 2005) of Mongolian sheep strains within a larger study of Chinese sheep populations. We couldn’t find study for Mongolia’s indigenous native sheep populations (as with domestic goats). Luo et al. (2005) found that there was totally same nucleotide composition among Mongolian and Chinese sheep mt-DNA D-Loop sequences. However, they reported slightly greater variation in Mongolian sheep (26.85% of the sites) than its Chinese sheep (24.22%). In generally, Mongolian sheep (11 strains) exists greater genetic diversity parameters than Chinese sheep (9 breeds), at number of haplotypes (86.06% (142/165) versus 78.83% (108/137)), haplotype diversity (Hd; 0.976 versus 0.936), nucleotide diversity (Pi; 0.036 versus 0.034) and a number of nucleotide differences (k; 23.50 versus 22.48) (Luo et al., 2005) using comparable sample size from each population.
Three haplogroups, A, B and C, were revealed in Eurasian sheep populations in previous studies (Hiendleder et al., 1998; Luo et al., 2005; Meadow et al., 2005; Chen et al., 2006). More recently, one more novel distinct lineage, D was described besides of A, B, and C by Tapio et al. (2006) and Liu et al. (2016). Lineage A was mainly found in Asian breeds, while lineage B was observed at the highest frequency from European breeds (Guo et al., 2005), while lineages C and D were found only in Asian breeds (Tapio et al., 2006). In case of Mongolian 11 strains, numerous number of animals from 9 of 11 studied populations were belonged to the lineage A (average frequency of 58.73%), few animals of 11 populations belonged to lineage B (with 24.68%), and the fewest animals from 10 out of 11 populations belonged to lineage C (with lower frequency 16.59%) (Luo et al., 2005). We found the same pattern for sheep as occurs for goats, in that Mongolia hosts 3 out of 4 lineages in its vast territory.
Genetic studies of domestic cattle, Bos taurus
The cattle are the most economically important domestic animals in the world, there are two main type of cattle are herding, namely beef cattle and dairy cattle (Lai et al., 2006). Previous studies suggested that they descended from aurochs Bos primigenius, species already extinct in several hundred years ago from Eurasian steppe (Payne and Hodges, 1997). The domestication event probably occurred 8,000-9,000 YB, and it tool place in several independent areas (Cai et al., 2007). In eastern Asia (including Korea and Mongolia), the domestic cattle may have been domesticated from local wild cattle or migrated from the early domestic center of the Near East (Mannen et al., 2004).
In Asia, totally 258 cattle breeds were reported, 11% of these breeds are recognized as being at the risk of extinction (Scherf, 2000). In addition to these breeds, native cattle in Asian fields, were not classified as breeds (Yonesaka et al., 2016). Historically, native cattle played an important role for Mongolian food and transportation demands since ancient times. Out of 1.57 million Mongolian cattle, 1.55 million supposedly belong to three indigenous Bos taurus cattle breeds which were divided and called as Mongol, Selenge and Khalkhun Golun. These breeds herd under broad pastoral systems. Mongolian native cattle generally have small body size, but sturdy and strong. The Mongolian breed is by the far the most common, with 1.53 million animals, and it is found throughout most of the country (Lkhagvaa et al., 2003).
We found a few previous studies on the population genetics of Mongolian cattle populations (Lkhagvaa et al., 2003; Mannen et al., 2003; Cai et al., 2007; Lin et al., 2010; Yonesaka et al., 2016). However, only few of these studies (Lhagvaa et al., 2003; Mannen et al., 2003; Lin et al., 2010) were mentioned for Mongolian cattle microsatellite (MS), mt-DNA and single nucleotide polymorphism (SNP) markers, respectively. Of this, heterozygosities (Ho; 0.679 and He; 0.639) in Mongolian cattle populations are similar to those obtained from northeast Asian taurine breeds (Ho; 0.647 and He; 0.660), but the heterozygosity values are higher compared to European (Ho; 0.598 and He; 0.577) and African taurine breeds (Lkhagvaa et al., 2003). The not close genetic relationship (data not shown) between studied three Mongolian cattle populations (North Hangai, Gobi-Altai and Khalkhun Golum; Lkhagvaa et al., 2003; Figure 2B) is suggesting that probably there are high level of crossbred with other introduced foreign breeds during Soviet Union period. In addition, Yonesaka et al. (2016) showed that Mongolian cattle have higher genetic diversity (Ho= 0.394, He= 0.389) than 15 other Asian cattle populations. They also found a close genetic relationship among Japanese, Korean and Mongolian cattle populations by an UPGMA tree, Principal Component and K=5 in STRUCTURE. This result suggests genetic admixtures among Japanese Holstein, Japanese Black cattle, Korean and Mongolian (Figure 2A).
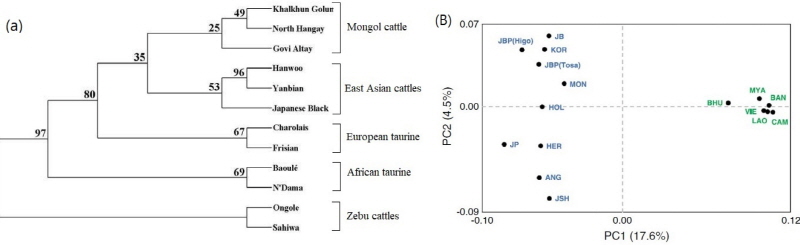
Figure 2 (a) UPGMA tree, constructed with the Reynolds’ Fst genetic distances, showing genetic relationships among the 12 cattle populations. Numbers indicated bootstraping values in percentage after 1,000 re-samplings. Sahiwal and Ongole were used as outgroups (data from Lkhagvaa et al., 2003). (b) Principal component analysis (PCA) of 16 cattle populations. Bos taurus population indicated by blue and Bos indicus by green letters (data from Yonesaka et al., 2016).
Mannen et al. (2004) investigated Korean and Mongolian cattle samples and found 23 and 32 distinct Bos Taurus haplotypes, respectively. Interestingly, the authors found that nine mt-DNA chromosomes in Mongolian cattle were discovered as the B. indicus type by comprised five mt-DNA haplotypes. This is probably due to the fact that Mongolia in the thirteenth century was one of the most vast empires in history and this conquest expanded to Southeast Asia, Southwest Asia and northern India (Rupen, 1979). Previous findings suggest that Mongolian livestock has a higher diversities in their both MS and mt-DNA markers due to the influx of foreign genetic resources during also Soviet Union period (same as other animals).
Genetic investigations into the improvement of Mongolian domestic animals
The agriculture sector is great important for the economy of Mongolia, and its share of the gross national product of the country exceed 14.6% in 2012 (and was 30% in the begining of the 2000s). The livestock sector produces more than 80% of the gross agriculture product (Shagdar 2002, Yadmaa 2014). As mentioned before, Mongolia reached 61.2 million head of domestic animals, and this tends continuously increase annually. Unfortunately, there is a lack of genetic investigation into this huge domestic animal population, which could negatively impact the development of an animal breeding program. In Mongolia, some breeding programs began in 1960s under the control and management of Russian scientists. Until the Soviet collapse in begining of 1990s, these animal breeding programs were rapidly developed including the introduction of some alien breeds from Soviet Union countries.
There are several reasons to increase and improve genetic investigations into the domestic animals of Mongolia. Improved genetic investigations could: (a) be a main driving force behind designing new breeding programs in Mongolia to improve the quality of livestock products for increased exportation, (b) help control the rapidly increasing population of domestic animals by producing higher quality animals, and (c) help control increases in disease among Mongolian domestic animal populations. However, these genetic improvement will take a time, and require additional genetic investigations. Most important things, the three genetic improvements could form the basis for future genetic studies of Mongolian domestic animals among foriegn and domestic researchers. Numerous challenges face improved genetic research: (i) lack of personnel and materials for genetic investigations, (ii) lack of or poor infrastructure and institutional facilities for field and lab work, (iii) herders’ poor knowledge of the importance of genetic improvements to their dometic animals, and (iv) lack of goverment funding arrangement for genetic research (indeed any basic research).
Conclusion
In conclusion, the valuable genetic resources exist in Mongolia’s domestic animals, including camels, horses, goats, sheep and cattle. Archeological evidence suggests that some domestic animals, especially horses and cattle may have been independently domesticated in Central Asia, including the Mongolian steppe. In addition, most previous population genetics studies used few samples (1 to 62) from Mongolian animals and only as a dataset for comparison, while only one or two studies fully explored the genetics of Mongolian domestic animals. These studies, especially on sheep and goats, used samples from common strains of these domestic animals, with no research into native Mongolian animals. According this review, we can also conclude that there were clear domestic animal migration ways from central Asia (Mongolia) through Korean peninsula to the Japan islands, especially for horse and cattle. Our review demonstrates a need for more genetic research into domestic animal populations in Mongolia to better understand their genetic importance. Finally, both the government and NGOs need to improve the quality and number of genetic investigations.
Further genetic research: We plan to conduct population genetics research into sheep, goats and cattle populations in Mongolia to contribute understanding of the genetic characteristics of Mongolian livestock using mitochondrial DNA genomic SNP markers. We will collect blood and tissue samples from central and, to an extent, southern Mongolia, representing Mongolian native sheep, goats, and cattle, including some breeds of sheep and goats that were not covered in previous studies.