INTRODUCTION
Intensive rearing of livestock at commercial scale is due to their consumption as main source of animal protein. Muscovy Ducks are among these poultry species because the steady increase of duck meat consumption; duck farming and production are currently receiving greater attention. Thus, recent interest in research and development of duck production is on the rise as well as breeding of duck. Continuous breeding of ducks needs basic information on their genetic background and diversity; because few reports are available about genetic information of Muscovy duck populations (Pingel, 1990; Knust et al. 1995; Romboli, 1995) any contribution on their genetic diversity worldwide will assist the breeder in large quantity and more viable duck production.
Recent advances in molecular genetics techniques has revealed genetic information of domestic animals with high accuracy compared to the information obtained by pedigree relationship and trait phenotypes. Such techniques have been successfully employed to address the genetic variation and, in turn, the genetic diversity among the different population which has been utilized in breeding program. Molecular markers and DNA sequencing have been taken as good markers to classify the taxonomy and phylogenetic relationships among species and the detailed genetic information of animals is available with high accuracy compared to the information obtained by pedigree relationship and trait phenotypes. The relationship between molecular techniques and conventional animal breeding methods to identify animals with higher genetic potential tends to maximize the genetic gain for the traits of interest (Schmidt et al., 2000), such techniques have been successfully employed to address the genetic variation and the genetic diversity among different populations and have been of great help in breeding programs. Few studies have been recently performed to assess the genetic polymorphism in ducks using a DNA fingerprinting technique. Maak et al. (2000) developed microsatellite markers for White Pekin and Muscovy ducks, Dolmatova et al. (2000) studied the possibility of using RAPD markers for the detection of differences among lines of White Pekin ducks. In addition, TianFang et al. (2002) studied the polymorphism in DNA bands for six duck breeds based on RAPD and reported high phylogenetic distance between Muscovy and other domesticated ducks. These are part of the evidences supporting further studies on Muscovy Ducks populations from Nigeria in order to establish the degree of genetic diversity. One of the recent study by Adeola et al. (2020) revealed low genetic diversity in Nigerian Muscovy duck based on mitochondrial D-loop. Studying Muscovy ducks genetic diversity could assist in breeding programme designed for their continuous production and prevention of inbreeding among the Muscovy duck population.
Therefore, this study was designed to analyze Nigerian Muscovy duck population with the aim of estimating their genetic diversity and relationship of Muscovy duck individuals from 15 different locations in four states in Nigeria using mtDNA CYP2UI genes.
MATERIALS AND METHODS
Animal sampling
A total of 82 Muscovy ducks were collected from 15 different locations in four states in Nigeria comprising of Kaduna, Kwara, Niger and Oyo. Blood sample was collected from each bird carefully through wing venipuncture. Total genomic DNA was extracted from ethanol preserved blood samples using a standard phenol-choroform method (Sambrook and Russell, 2001). Quality and quantity of the genomic DNA was checked by Agarose electrophoresis and Nanodrop respectively.
Polymerase chain reaction and DNA sequencing
The 747 bp nuclear DNA CYP2UI gene fragment was amplified using the following primers P103 (5-GTTATTTGGTTAGCATATCGTG-3) and (5-GAGACGGTTGCGTATATGG-3). PCR amplification was carried out on a thermocycler with a 25 ul volume reaction mixture, which included 50 ng of genomic DNA, 12.5 ul of master mix (Promega, Fitchburg, WI) and 2 ul of (10 pmol) of each primer. The PCR was perfumed at 95℃ for 5 minutes, follow by 30 cycles of 30 seconds at 94℃, 1 minute at respective annealing temperature (cytochrome; CY2UI:45℃) and 1min at 72℃. The amplified PCR products as sequenced using the Big Dye terminator cycle sequencing kit. (Applied Biosystems, Foster City, CA) on ABI 3500 Genetic Analyzer.
Data analysis
The CYP2UI gene sequence was manually edited aligned and an un rooted neighbor-joining (NJ) typology was constructed drawn using Clustal W implemented in MEGA 7.0.18. (Tiamura et al., 2016). Genetic diversity indices such as a number of haplotypes, haplotype diversity, polymorphic sites and nucleotides diversity were analyzed using Dna SP. v. 6.12.03 (Rozas, 2009). The sequences were further subjected to Multiple Sequence Alignment MSA viewer using Clustal x 2.0 and Jalview 1.8.3. (Larkin et al.,2007). A median-joining network (Bandelt et al., 1999) was constructed with network 10.2 ().
RESULTS
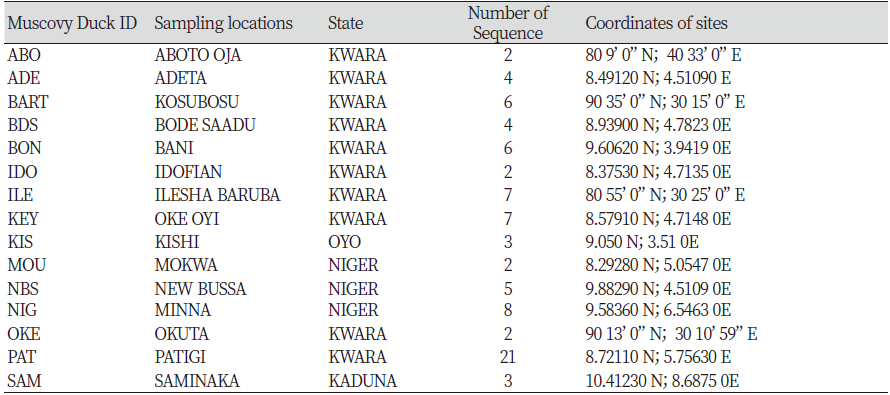
Table 1 showed the Muscovy Duck ID, sampled locations; number of ducks sequence obtained and coordinates of the sampling sites. A total of 82 sequences were obtained across the 15 locations out of which one location each was sampled from Kaduna and Oyo state, 10 locations from Kwara State, and three from Niger (Figure 1).
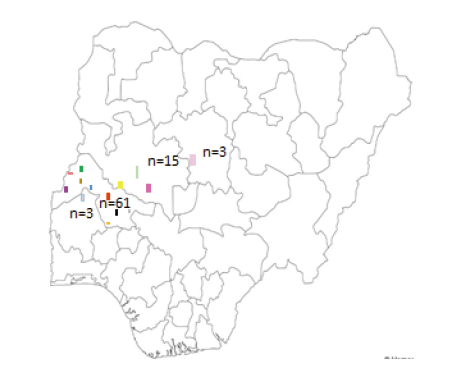
Figure 1. Proportion of individuals sampled from each region with colour indicating the sampling in Kaduna, Kwara, Niger and Oyo States of Nigeria: Blue-ABO; Black-ADE; purple-BART; Red-BDS; Brown-BON; Orange-IDO; White-ILE; Grey-KEY; Light Blue-KIS; Yellow-MOU; Pink-NBS; Lime-NIG; Light Red-OKE; Green-PAT; Light Pink-SAM.
Table 1. Muscovy ducks ID, Sampling locations, Obtained sequence and coordinates of the Sampling Sites
|
Total Sequence Variation, Haplotype and Nucleotide diversity
Eighty two sequences were obtained (61, and 15 sequences from Kwara and Niger state, respectively; 3 from Kaduna and Oyo State). The total length of sequence studied is 747 bp with a total of 48 variables sites or approximately 6.43 of the full length. Figure 2 showed some polymorphism (substitutions) in the aligned sequences. The polymorphic sites consist of substitutions with 33 singletons variables of two, three and four variants with a value of 30, 2 and 1 respectively, (Table 2).The Nucleotide diversity per site (Pi ) was 0.00587, while total number of mutation was 52 and the parsimony informative site was 15 as shown in Table 2.
Table 2. Total Sequence Variations and Haplotype diversity between the population of 82 Muscovy Ducks Sampled

|
Phylogenetic analysis
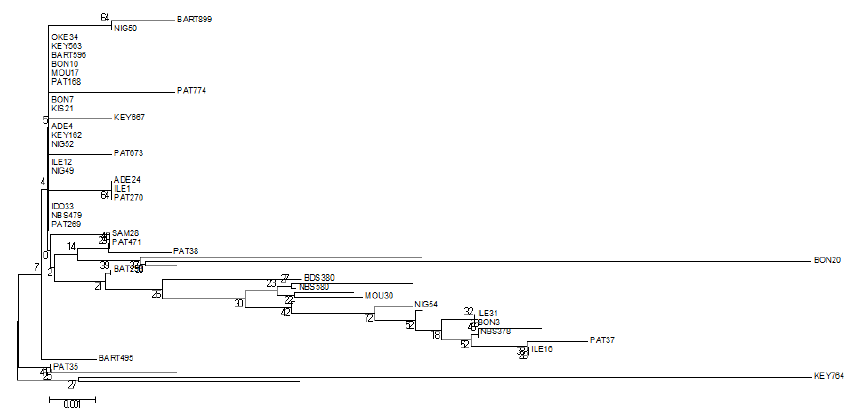
Network of 82 domesticated Muscovy ducks samples constructed using NETWORK v. 10.2 showed the connectivity between some population clustered together at the epicentre, with few far apart in a distant relationship (Figure 2). The phylogentic tree also support a closer relationship between the Muscovy ducks except PAT 35 and KEY 764 (Figure 3).
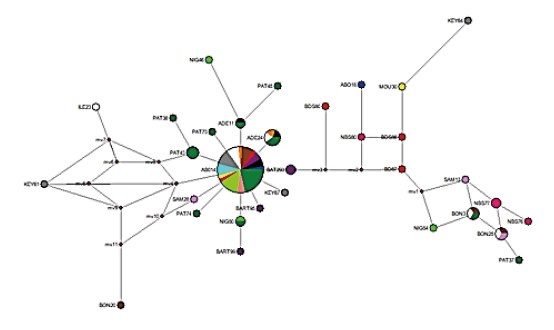
Figure 2. Median-joining network of 82 Nigerian Muscovy duck samples constructed by using network 10.2. Colours indicate the geographical distribution of the sampling locations across States in Kaduna, Kwara, Niger and Oyo. Blue – ABO (ABOTO); Black – ADE (ADETA); Purple – BART (KOSUBOSU); Red – BDS (BODESAADU); Brown – BON (BANI); Orange – IDO (IDOFIAN); White – ILE (ILESHA BARUBA); Grey – KEY (OKEOYI); Light Blue – KIS (KISHI); Yellow – MOU (MOKWA); Pink – NBS (NEW BUSA); Lime – NIG (MINNA); Light Red – OKE (OKUTA); Green – PAT (PATIGI); Light Pink – SAM (SAMINAKA).
Haplotype and Nucleotide diversity within the Sampling Locations.
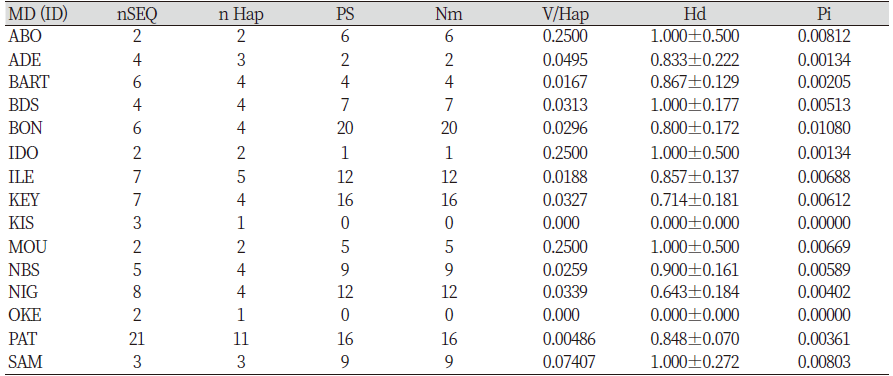
Table 3 showed the haplotype diversity of ABO, BDS, IDO, MOU and SAM to be 1; 0 Hd was recorded in KIS and OKE, NBS had highest Hd 0.900, those of ADE, BART, BON, ILE, and PAT were high and ranged from 0.800 to 0.867; KEY and NIG also had high haplotype diversity of 0.714 and 0.643, respectively. The overall haplotype of all the sequence ranged from 0 to 0.900, with haplotype number of 1 to 11 and polymorphism site of 0 to 20. The variance of haplotype diversity ranged from 0 to 0.25 while the nucleotide diversity ranged from 0.00 to 0.00812. PAT had the highest sequence of 21 with 11 haplotypes, while BON had the highest polymorphism site (20). KIS and OKE had no polymorphism. BON had the highest mutation (20) followed by PAT and KEY (16), no mutation were recorded in the sequences of KIS and OKE. (Table 3).
DISCUSSIONS
Muscovy duck lovers in Nigeria have been raising this species of poultry at their backyard for ages for consumptions, income, companionships, entertainment and ritual purposes. Muscovy ducks are regarded as valuable birds and attract higher prices than other indigenous poultry species in the market (Sola-Ojo et al., 2020). Rearing of Muscovy ducks are not devoid of Migration in the county and this has introduced a long distance gene flow and enables genetic materials exchange within the commonly available species of Muscovy ducks and other poultry species ( Liu et al., 2006; Cuc et al., 2011).
Data obtained for genetic variations are useful in genetic diversity within and between populations because migration and population history infers population genetics parameters, they are also useful in studying the mechanisms of nucleotide changes and estimation of recombination rate (Hammer et al., 2004). Studying the genetic background and relationships among populations of Muscovy duck will provide information on the relationship within their local family, between locations interactions ad relationships, ancestral lineage, the rate of genetic diversity within and between different population and could assist the breeder by providing useful information for a planned breeding programme as ducks owners practiced random mating in Nigeria. This study provides molecular genetic characteristics, and diversity among Muscovy duck found in few selected state closer to and within North central Nigeria.
As shown in Table 2, the Muscovy duck found in the 15 locations of studies were distributed into 32 haplotypes with haplotypes diversity of 79.25%, which is an indication of existence of contribution of multiple maternal lineage across the location and this correspond with the findings of Liu et al. (2004); Hoque et al. (2013); Liao et al. (2016) where maternal lineage sharing were reported among different indigenous chickens of various geographical locations.
Similar sequences expressed as the same haplotype of Muscovy duck for different locations in this study suggest that Muscovy duck are in close genetic relationships irrespective of distinctive and varying phenotypic, biochemical and some physiological characteristics as reported by Sola-Ojo et al. (2020). Hybridization among the Muscovy duck varieties in this study with regards to different locations could be responsible for the rate of haplotype diversity between the populations as asserted by Piyanat et al. (2018) for Thai chickens. No mutations observed in sequences from OKE and KIS could be as a result of small sample size.
In this study, the phylogenetic tree revealed a clustered relationship and this corroborates the fact that domestic animals migrated from one location to the other through human activities and there are greater tendencies for intensive genetic admixture as stated by Adeola et al. (2015). Also, Nigerian Muscovy duck have been managed and raised with no planned breeding programme thus the impacts of selection pressures will be smaller and may not applied to their population. However, a distant genetic relationship were observed in PAT 35 and KEY 764, and this might be as a results of migration or random genetic drift in their natural populations
In conclusions, this study showed that Nigeria Muscovy duck are likely a descent of common maternal lineage under genetic equilibrium within and between genetic variations across the studies locations.