INTRODUCTION
Nigerian local chicken (NLC) is widely kept across Nigeria constituting 80% of the National poultry flock (60% of 120 million chickens). Though low in growth rate relative to exotic chicken, NLC are favoured with exceptional qualities (including hardiness, biotic and abiotic stress tolerance, gustatory quality of meat, high brooding efficiency, high fertility and hatchability) and preserved for the same reason (RIM, 1992). Marker assisted selection offers a highly attractive, efficient and effective tool for improving growth performance of the NLC while preserving its existing merits. To use Marker Assisted selection, polymorphisms within genes known or presumed to determine variation in the trait of interest (or to be closely linked) must first be identified. Subsequently, linkage or association studies in populations in which the trait of interest and known polymorphisms in the gene are segregating are used to test whether and to what extent the gene of interest co-segregates with the trait of interest and by implication is causally linked or associated. Linked/associated markers are then used to select birds that carry the allele associated with superior trait performance in the straight-bred population under improvement. Equally, where the superior allele is absent from a breed under improvement interest, crosses may be made between a breed carrying the allele of interest and the breed under improvement, followed by iterative crossing and selection of birds carrying the superior allele in each generation to introgress the marker’s linked to superior trait performance with speed and efficiency. This will improve supply and contribute to animal protein sufficiency in Africa.
Several candidate gene determinants of growth may be amenable to selection but such an approach is currently not possible because polymorphisms that may contribute to candidate gene function in determining growth in the NLC have not yet been identified to fill the existing gap in knowledge.
The present study was designed to examine the nucleotide sequence of the gene that encodes chicken Fatty Acid Binding Protein 4 (FABP4) also known as Adipocyte Fatty Acid Binding Protein (AFABP), a growth associated gene, for polymorphism that may determine differential protein function between fast growing broiler chickens (Hubbard breed) and slow growing Nigerian Local Chicken (Fulani Ecotype) as a prelude to marker confirmation and use in improvement of the latter. Earlier work by others did implicate FABP4 in cell growth and differentiation, cellular signaling, gene transcription, and protection of enzymes from the toxic effects of free fatty acids (Glatz and Veerkamp, 1985; Zimmerman and Veerkamp, 2002), which are all component of growth. Fatty acid-binding proteins (FABPs) such as A-FABP/FABP4 exhibit a high affinity for long-chain fatty acids and appear to function in the metabolism and intracellular transportation of lipids in poultry birds. According to Fisher et al. (2001), FABP4 is expressed in many tissues, especially in adipose tissue, and is associated with lipid metabolic enzymes. The FABP4 gene is also regarded as a candidate gene for intramuscular fat (IMF) deposition, as it can enhance the deposition of triglyceride during adipocyte differentiation in muscle (Gerbens et al., 1998; de Koning et al., 1999; Saez et al., 2009). Intramuscular fat which contribute to overall tissue weight has been described to have a positive influence on meat quality, with respect to tenderness and flavor of the meat (Wood et al., 1999; Mourot and Hermier, 2001). Because polymorphisms that disrupt gene function are more likely to be found in the conserved portion of the genome, including regulatory and coding portions of genes (promoters, splice cites, and exons), the study targeted the complete collection of exons constituting the coding portion of the gene (i,e, the FABP4 exon) and the 50 base pair non-coding region immediately preceding (5’) and following (3’) each exon, in which splice donor and acceptor sites reside and in which other regulatory sequences may also reside.
The present study was predicated on three linked hypotheses; that differences in FABP4 gene coding and near-adjacent DNA sequences exist between slow (Fulani NLC) and fast growing (Hubbard broiler) chickens; that through in silico translation of DNA variant/ polymorphic forms distinguishing between the examined groups, consequences of polymorphism at the level of primary protein structure (peptide sequence can be ascertained), and; through comparison of the region of polymorphism in chickens against genetically diverse extant species, clues of the evolutionary constraints on variation of the polymorphic region (which point to functional importance of such a region) can be gleaned.
MATERIALS AND METHODS
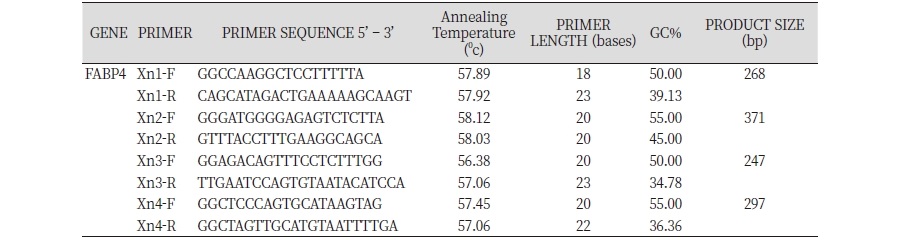
Primer Design
The sequence of the primary transcript of FABP4 gene (ENSGALT00000025427.3) of Red Jungle fowl was retrieved from Ensembl (). Forward and reverse primers (18 – 23 base length and 58 ± 50C melting temperature) were designed to amplify each exon and the 50 base sequences immediately preceding and following each exon and synthesised through a commercial service (Inqaba Biotech) and used to amplify their target region in Hubbard Broiler and FE-NLC genomic DNA by use of the Polymerase Chain Reaction (PCR).
Blood Collection and DNA Extraction
Blood samples were obtained from Hubbard and Fulani ecotype. Briefly 1ml blood was drawm from the brachial vein through a 23 gauge needle into a 2ml syringe and aspirated in a tube containing ethylene diamine-tetra acetate (EDTA)
DNA was extracted from each sample by use of the procedure of Wang et al. (2006). Yield and purity of DNA samples were determined using Nano-drop (NANODROP 2000) spectrophotometer.
Amplification of Exons
A PCR cocktail containing 12.5μl New England Biolabs 2X Master mix (20mM of Tris-HCl, 1.8mM of MgCl2, 22mM of NH2Cl, 22mM of KCl, 0.2mM of dNTPs, 5% glycerol, 0.06% IGEPAL, 0.05% OF Tween, Xylene Cyanol FF, Tartrazine and 5 units of Taq DNA polymerase), 4μl of 25ng/ μl chicken DNA template, 2.5 μl (25pM) each of forward and reverse primer and distilled deionized H2O to a final volume of 25μl. PCR reactions were performed in an MG96G MyGene (LongGene Scientific Instrument Co.,Ltd.) peltier effect thermal cycler, programmed for 2’ at 95°C, followed by 35 cycles of denaturation (30" at 95°C), annealing (1’ at 55°C, 58°C or 60°C) and extension (30" at 72°C) and a final step of final incubation of 3’ at 72°C. Amplicons were resolved by gel electrophoresis (1% agarose, 0.5 µg /ml ethidium bromide 10V/cm, 45’). Stained gels were view under a UV trans-illuminator.
Sequencing
Amplicons products were subjected to forward and reverse primer sequencing on an ABI 3500XL Genetic Analyzer (Applied Biosystem Inc.) through a commercial service (Inqaba Biotechnology Laboratories, South Africa). Output files were viewed using Finch TV v1.4.0. (Geospiza, PerkinElmer, USA).
Multiple Sequence Alignment
Multiple Sequence Alignment was performed by use of MultAlin (Corpet, 1996). Each sequence was first aligned against the predicted transcript from the chicken genome sequence derived from Red Jungle Fowl (ENSGALT00000025427.3) to verify that the intended portion of the target gene was correctly amplified and sequenced.
In silico
Sequences were translated in-silico using a standard genetic code by means of ExPASy Translate tool (Swiss Institute of Bioinformatics, ).
Phylogenetic Analysis
Orthologs of chicken FABP4 (from order aves, eutherian and non-eutherian mammals) were retrieved from public databases () and used in construction of phylogenetic tree using Phylogeny.fr (Dereeper et al., 2008).
RESULTS
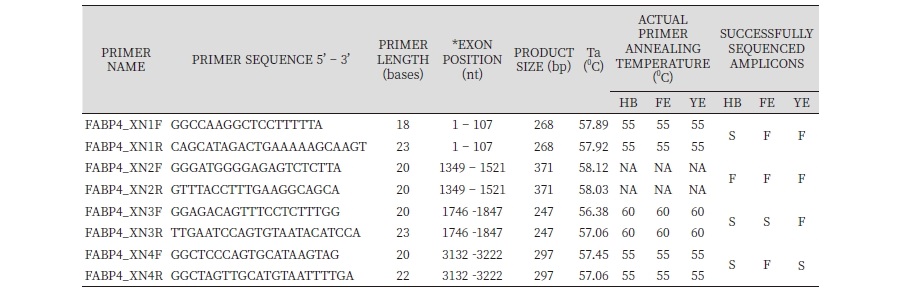
Three out of four primer pairs corresponding to FABP4 exon 1 to 4 (Table 2) successfully amplified a fragment of the predicted size in at least one of the three breeds examined (Hubbard broiler, Fulani Ecotype and Yoruba Ecotype NLC) although non-amplified fragments in all three breeds as shown in Table 2 and Figure 1
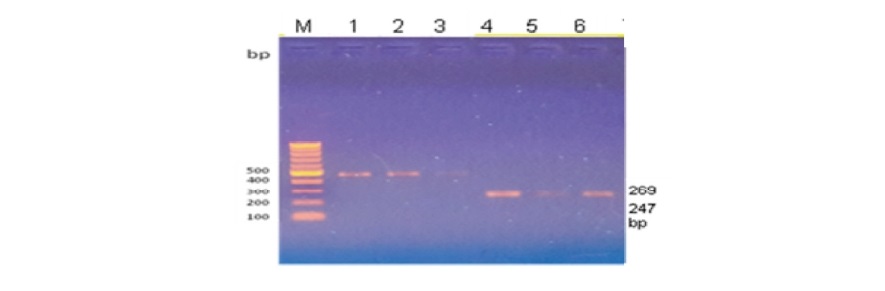
Figure 1. Agarose gel resolved fragment of PCR FABP4 product showing (left – right column), Lane M = Marker (100bp DNA ladder), lane 4, 5, and 6 represent Fulani Ecotype (FE), Yoruba Ecotype (YE), Hubbard Broiler (HB) FABP4 exon 3 respectively, fragment sizes of the marker are listed on the right side.
DNA Primary Structure – Nucleotide Sequence
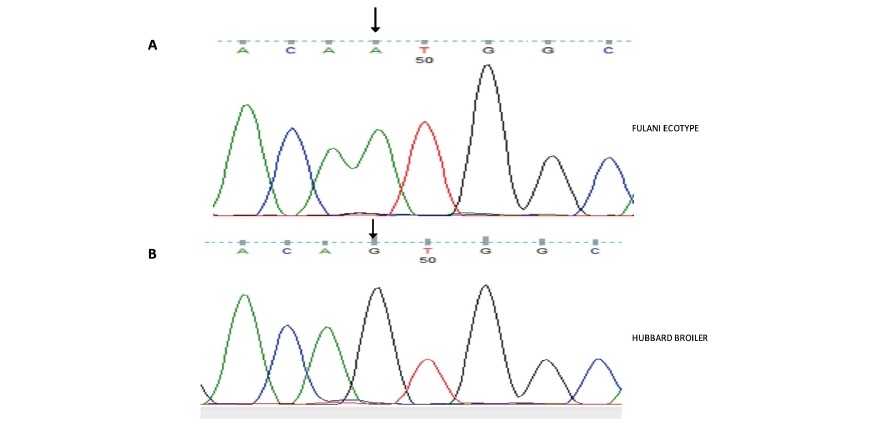
Multiple sequence alignment of the Fulani ecotype, Yoruba ecotype and Hubbard Broiler FABP4 exon 3 sequences and flanking intron DNA produced in the present study against exon 3 of the Red Jungle Fowl primary transcript (ENSGALT00000025427.3 nt 1741 – 2067) revealed the existence of a purine to purine Single Nucleotide Polymorphism (SNP) in position 1765 (i.e. position 20 of exon 3; position 266 of the CDS) as shown in Figure 2. The Red Jungle Fowl and Fulani Ecotype chicken carried an Adenine (A) base and broiler carried Guanine (G) (c. 266A>G) as shown in Figures 2 and 3.
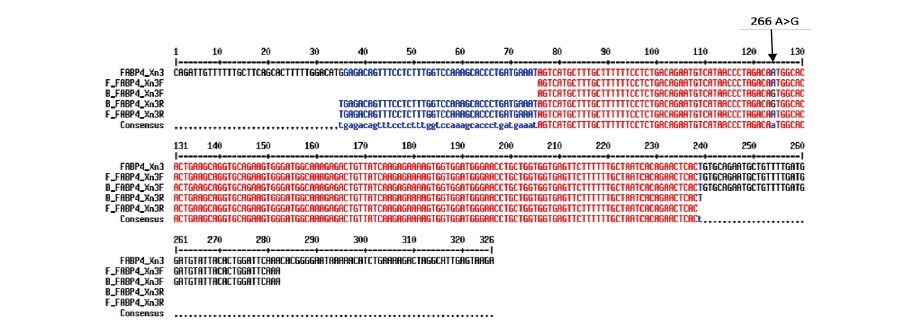
Figure 2. Multiple DNA sequence alignment of FABP4 exon 3 from; Red Jungle Fowl ENSGALT00000025427.3 nt 1741 – 2067 (FABP4_Xn3); Fulani, using forward (F_FABP4_Xn3F) and reverse (F_FABP4_XnR) primers in sequencing and; Hubbard Broiler using forward (B_FABP4_Xn3F) and reverse (B_FABP4_Xn3R) primers in sequencing. Polymorphic base shown (arrow).
Predicted peptide sequence
Multiple Sequence Alignment of Peptide sequences from the translation of Jungle Fowl (ENSGALT00000025427.3 nt 1741 – 2067), Fulani and Hubbard broiler sequences (Figure 4) showed that the c. 266A>G mutation resulted in a missense variation at codon 7 of exon 3 (codon 89 of the full peptide sequence), in which codon 7 of exon 3 in Red Jungle fowl encoded the polar amino acid Asparagine (Asp, N), and the corresponding codon in broiler encoded Serine (Ser, S) (also a polar amino acid), as shown in Figure 4 i.e. N7S polymorphism.
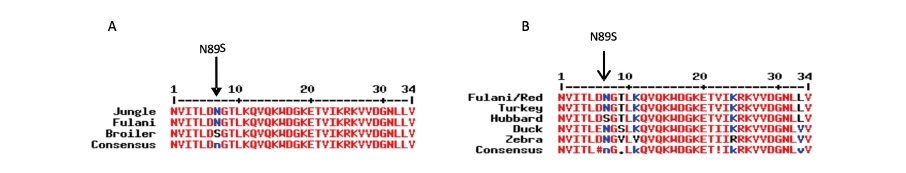
Figure 4. (A) Alignment of peptide sequence of Red Jungle Fowl (ENSGALP00000025381 Codon 83 – 116) FABP4 exon 3 and corresponding peptides from in-silico translated Fulani ecotype and Hubbard broiler gene sequence. Polymorphic Peptide shown (arrow) at codon 7 (of exon 3) corresponding to codon 89 of the full length of FABP4 CDS. (B) Alignment of the FABP4 exon 3 peptide sequences of Fulani ecotype/Red Jungle Fowl, Turkey, Hubbard Broiler, Duck and Zebra Finch, showing polymorphic peptide at codon 7 of Exon 3 of FABP4. Polymorphic base shown (arrow).
Multiple sequence alignment of the predicted peptide sequence of FABP4 exon 3 from three randomly chosen species representing three genera within the taxonomic order Aves (Turkey, Duck and Zebra finch) along with predicted peptide sequence of exon 3 of Broiler and Red Jungle Fowl/Fulani chicken (Figure 4B), revealed that the use of asparagine at codon 7 of exon 3 in the Jungle Fowl/Fulani chicken is conserved in avian evolution
Multiple peptide sequence alignment of FABP4, exon 3 for Aves, Eutherian and non-eutherian mammals showed that FABP4 exon 3 codon 7 (numbering relative to the chicken exon) was less well conserved over a broad evolutionary history spanning over 300 million years captured in the set of exon 3 peptide sequences examined (Figure 5).
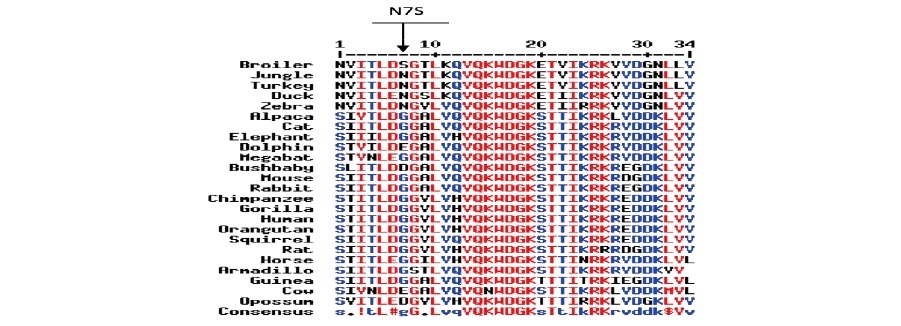
Figure 5. Multiple peptide sequence alignment of FABP4 exon 3 for Aves, Eutherian and non-eutherian mammals by use of MultAlin software (Corpet, 1996), showing the N7S polymorphisms. Peptides numbered relative to Red Jungle Fowl FABP4 exon 3 peptide sequence. Chicken (ENSGALP00000025381), Alpaca (ENSVPAP00000001054), Duck (ENSAPLP00000005715), Armadillo (ENSDNOP00000033224), Bushbaby (ENSOGAP00000011712), Cat (ENSFCAP00000017433), Chimpanzee (ENSPTRP00000034857), Cow (ENSBTAP00000000079), Dolphin (ENSTTRP00000006583), Elephant (ENSLAFP00000000507), Gorilla (ENSGOPP00000007182), Guinea pig (ENSCPOP00000006483), Horse (ENSECAP00000021250), Human (ENSP00000256104), Megabat (ENSPVAP00000013471), Mouse (ENSMUSP00000029041), Opossum (ENSMODP00000008038), Orangutan (ENSPPYP00000020978), Rabbit (ENSOCUP00000013299), Rat (ENSRNOP00000073333), Squirrel (ENSSTOP00000007662), Turkey (ENSMGAP00000012217), and Zebra Finch (ENSTGUP00000012023).
Protein Prototype Assisted Modelling
Comparative Protein prototype assisted modelling of the folded/tertiary/3-dimensional structure of FABP4 containing the peptide Serine at position 89 (Hubbard broiler) and the alternative protein structure containing Asparagine at position 89 (Fulani/ Red Jungle Fowl) of the complete FABP4 CDS revealed no profound effect of the N89S missense mutation on protein structure (Figure 6).
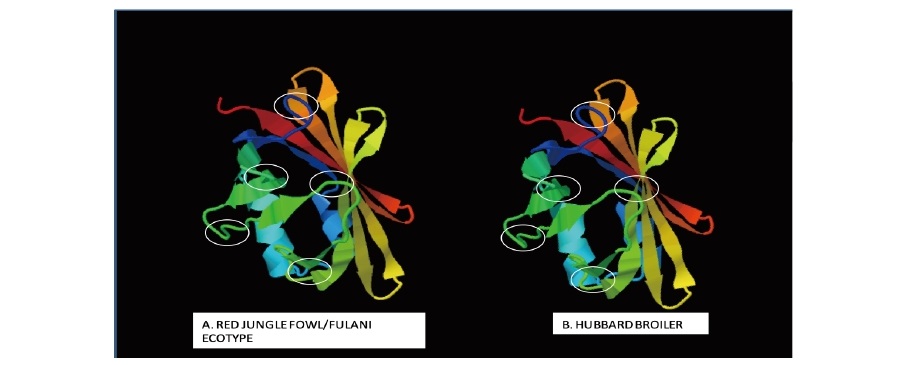
Figure 6. Predicted 3D structure of FABP4 protein encoded by Red Jungle Fowl/Fulani genome carrying the exon 3 peptide variant N (A) and structure of the corresponding Hubbard Broiler protein carrying the exon 3 peptide 7 variant S (B). Structure model produced by Phyre 2 software. White rings in the corresponding areas of A and B highlight structural conformational differences between the two models.
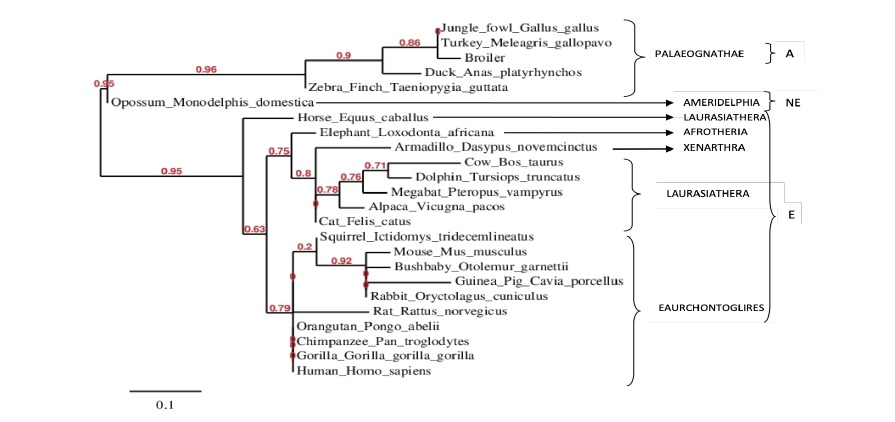
Phylogenetic Analysis of FABP4 Exon 3
A phylogram constructed from FABP4 exon 3 peptide sequence of chicken exon 3 of FABP4 of Hubbard Broiler, Red Jungle Fowl and Fulani Ecotype chicken, and other Aves (duck, turkey, zebra finch) in the Palaeognathae super order, using Eutherian and non-eutherian mammals as out-group, showed that the Red Jungle Fowl and Fulani Ecotype allele N89 (c. 266A) is ancestral to the broiler allele S89 (c. 266G) and the broiler allele is the more recently evolved form, the conventions c. 266A>G and N89S are therefore most appropriate in describing the mutation at the level of DNA and peptide sequence as shown in Figure 2 and 3 and Figure 4B and 5 respectively.
DISCUSSION
The Adenine (A) to Guanine (G) Single Nucleotide Polymorphism (SNP) at position 266 of the mature RNA/ position 20 of exon 3 of the FABP4 gene identified in the present study had previously been described (SNP Identification number rs431850290) by Luo et al. (2006) and Wang et al. (2009) in independent studies on Arbor Acres broiler chickens, and that the Red Jungle Fowl A266 variant was carried by the Baier egg laying Chinese Local Chicken breed. It was suggested that the N89S mutation allele might be associated with higher fat deposition (Wang et al., 2006). Zheng et al. (2009) found that FABP4 was more expressed in fast growing birds than in slow growing birds whereas a fat breakdown related enzyme thiolesterase B is expressed higher in slow growing birds, suggested that different metabolic regulatory network are indispensable for differential growth rate between fast and slow growing bird.
A Cytosine (C) to Thymine (T) substitution at the DNA level which is a silent (synonymous) mutation ( as it did not change the function of the amino acid as both still encoded phenylalanine) was also previously detected by Wang et al. (2006) in a separate study in exon 1 of chicken FABP4 using China Agricultural University Resource Population (CAURP) and Northeast Agricultural University Resource Population (NEAURP) which was obtained by crossing broiler sires, derived from lines at NEAU divergently selected for abdominal fat, with Baier layer dams, a Chinese local breed. The C variant is ascribed to Baier (local Chinese egg layer) grandsire and the T is ascribed to Broiler grandsire (Arbor acre). This may be due to linkage disequilibrum between the polymorphic base and a proximal or distal causal variant and/or linkage diequilibrum between other loci acting in Trans.
A study of the same location in the present work revealed no variation was found at the revealed base position for Hubbard broiler and Yoruba ecotype.
Luo et al. (2006) and recently reported separately that the expression of FABP4 significantly correlated with abdominal fat, breast intramuscular fat and thigh intramuscular fat and was highly expressed in the Beijing-YOU chicken compared to the Jing Xing chicken.
Although it was unclear to the author whether the exon 3 A to G substitution in the chicken FABP4 gene provides a molecular explanation for the previously reported linkage between a silent mutation in exon 1 of the gene and chicken abdominal fat, subcutaneous fat and intramuscular fat contents earlier reported by (; ), it seems to us the most parsimonious explanation for their results.
In the present study, multiple sequence alignment using Aves (Broiler, Jungle Fowl/Fulani, Turkey, Duck and Zebra finch) as in group and with peptide sequences which showed that the use of asparagine at codon 7 of exon 3 (amino acid position 89 of the FABP4 coding sequence) is conserved in avian evolution, showed that, while at the nucleotide level the SNP represents a purine to purine change, its consequence at the amino acid level is a substitution of a polar for another polar amino acid. Its (the SNP) localization within an exon (coding region of the gene), its non-synonymous consequence at the level of amino acid/peptide sequence, and the difference in the properties of the alternative variant amino acids (asparagine and serine), coupled with the evolutionary conserved location of the non-synonymous substitution suggests that, it affects the function of the encoded protein (FABP4). Indeed, a search through the literature confirmed that others have shown previously that the asparagine to serine mutation alters the functional properties of the protein (Wang et al., 2009). The phylogeny of exon 3 for Palaeognathae was consistent with published literature on the molecular classification and phylogeny of mammals (Tarver et al., 2016). In the out-group however, although the molecular phylogeny of exon 3 was largely consistent with the overall phylogeny of mammals (Tarver et al., 2016), the super order Laurasiatheria was split with a suggestion that perissodactyla (a member of the super-order Laurasiatheria) split from Laurasiatheria prior to the emergence of super-orders Laurasiatheria, Xenarthra, Afrotheria and Eaurcongtoglires from a common ancestor. Relationships among orders of the eutherian and the non-eutherian mammals according to literature are largely unresolved partly because of their extreme rapid diversification in the late Cretaceous and early tertiary period and partly because of extremes in convergent and divergent adaptation to a wide range of habitats (Tarver et al., 2016). Nevertheless, the most parsimonious explanation for this anomaly is that Perrisodactyla is exceptionally conserved through evolution, and, that the horse has retained the ancestral form of exon 3 of FABP4 that predates the divergence of Laurasiatheria (to which it belongs) from the Afrotheria and Xenarthra.
The evolutionary and functional basis for differential (extreme) conservation of exon 3 of horse relative to other Laurasiatheria is unclear but may have implications for a differential function of horse FABP4 protein relative to orthologs for other Laurasiatheria. Exon 3 is more conserved across the primate (relative to the phylogram derived from alignment of the entire gene) suggesting functional conservation of exon 3 in this order (Primates), this was consistent with the findings of Tarver et al. (2016).
Previous studies (Hedges and Maxson 1996; Nishihara, et al., 2009; Wildman, et al., 2007) have suggested a clear pattern of biogeographic diversification for placentals into four principal lineages (Afrotheria, Xenarthra, Laurasiatheria and Euarchontoglires) caused by drift vicariance.
It can be concluded from this experiment that; Natural variation exists in the coding protein of FABP4 gene distinguishing exotic Hubbard Broiler from indigenous local chicken (YE-NLC and FE-NLC), and this results in change in the peptide sequence (primary structure at the level of sequence) of the encoded protein, with attendant consequence for function
The FABP4 exon 3 gene can as serve as a marker for function of the gene and its region of linkage disequilibrum in differential growth of Hubbard Broiler and Nigerian local chicken. The FABP4 exon 3 mutation (266A > G, N89S) may determine variation in growth rate, fat deposition and meat quality between exotic Hubbard Broiler and Nigerian Local chicken.
RECOMMENDATIONS
This Single Nucleotide Polymorphism (SNP) c 266 A >G should be used as a marker for studying effect of FABP4 on growth and performance in chickens.
Association studies should be conducted in a cohort (Hubbard broiler, Fulani ecotype, unimproved Yoruba ecotype and improved Yoruba ecotype) in which alleles are segregating.
The present study should be repeated to ensure sequencing of all exons of both (FABP4) gene for Hubbard broiler, Fulani ecotype and Yoruba ecotype so as to examine the consequence of variation in as-yet-unsequenced portions of the gene.